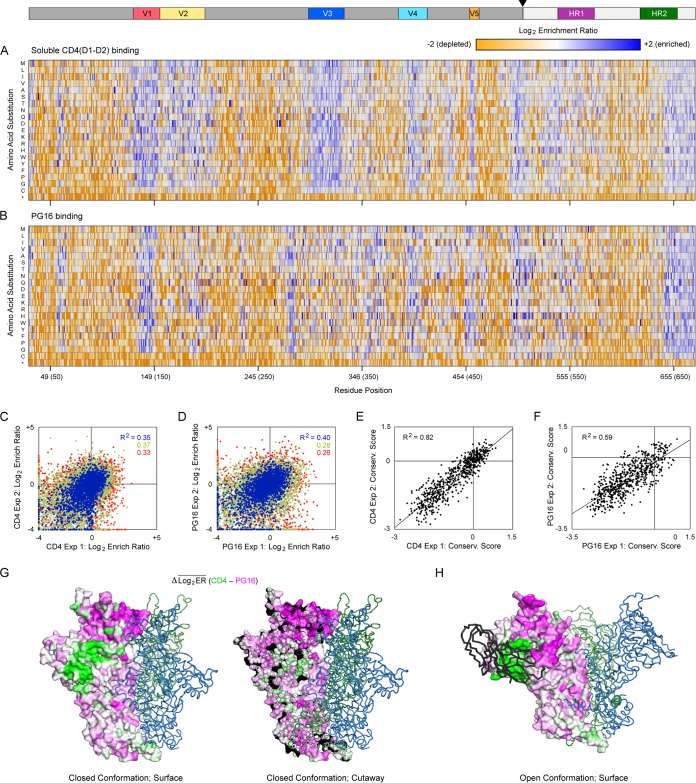
FIG 7.
Residues at the EnvDU422 trimer interface are more conserved for PG16 binding than for CD4 interactions. (A) SSM libraries of membrane-anchored gp140DU422 were expressed in Expi293F cells and sorted for binding to sCD4. Log2 enrichment ratios are plotted from ≤–2 (depleted, orange) to ≥+2 (enriched, dark blue). The primary structure of gp140 is on the horizontal axis, and amino acid substitutions are on the vertical axis. *, Stop codons. In the upper schematic, gp120 and gp41 are dark and pale gray, respectively; the cleavage site is indicated with an arrowhead; and notable regions are colored. Averages of two independent experiments are shown. (B) Mutational landscape of gp140DU422 under FACS-based selection for binding to PG16. (C and D) FACS-based selections for 10 nM sCD4 (C) and 3 nM PG16 (D) binding were independently replicated. Agreement between the replicate log2 enrichment ratios for each mutation is plotted. Abundant mutations (frequencies > 2 × 10−4 in the naive library) are blue, mutations with moderate representation (frequencies between 5 × 10−5 and 2 × 10−4) are green, and rare mutations (frequencies < 5 × 10−5) are red. (E and F) Agreement between the residue conservation scores from replicate selections for sCD4 (E) or PG16 (F) binding. (G) An atomic model of trimeric DU422 gp140 in the closed conformation, with one protomer shown as a surface, and the other protomers shown as dark green and blue ribbons. The PG16-CD4 conservation difference scores are mapped to the protomer surface in the same orientation as Fig. 2, with magenta indicating residues preferentially conserved for PG16 binding and green indicating residues more conserved for CD4 binding. On the right is a cross-section showing that preferential conservation for PG16 binding extends into the core of the trimerization domain. (H) DU422 gp140 modeled in the open state bound to CD4 (black ribbon; CD4 is shown bound to only a single protomer).