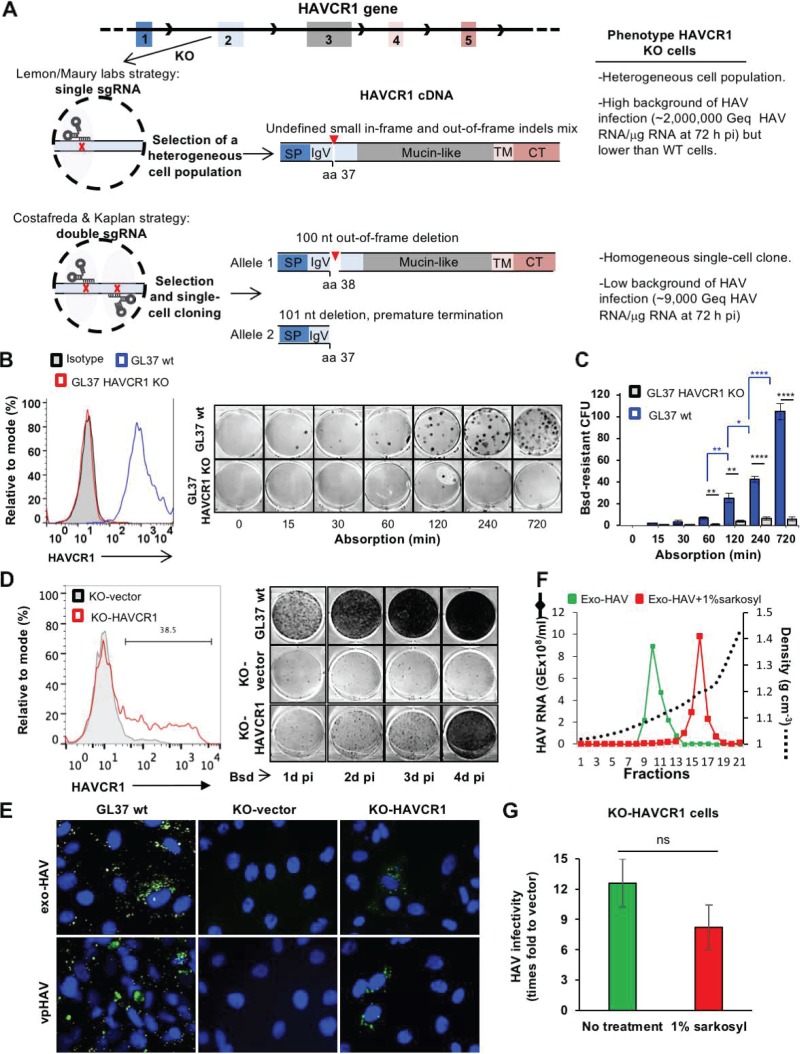
FIG 1.
(A) Schematic representation of the CRISPR/Cas9 strategies used to knock out the HAVCR1 gene at exon 2 in GL37 cells. The strategy used by Das et al. (11) (Lemon/Maury labs) was based on a single sgRNA to introduce a single cut in exon 2 and generate small and undefined indels. A heterogeneous population of cells containing indels was used to assess the role of HAVCR1 as a functional HAV receptor. The strategy adopted by Costafreda and Kaplan (1) was based on two sgRNAs that cut exon 2 at two different sites, resulting in defined out-of-frame deletions of 100 nucleotides in one allele and 101 nucleotides in the other allele of the selected single-cell clone used to assess the role of HAVCR1 as a functional HAV receptor. The HAVCR1 cDNA codes for a type I integral membrane glycoprotein containing a signal peptide (SP), a variable immunoglobulin-like domain (IgV), a mucin-like domain, a transmembrane domain (TM), and a cytoplasmic tail (CT). aa, amino acid; Geq, genome equivalents; pi, postinfection; nt, nucleotide. (B) Adsorption kinetics of vpHAV in GL37 wt and GL37 HAVCR1 KO cells. (Left) Analysis of the expression of HAVCR1 at the cell surface of GL37 wt (blue line) and GL37 HAVCR1 KO (red line) cells by flow cytometry analysis using anti-HAVCR1 MAb 1D12. GL37 wt cells were also stained with an isotype control (filled gray curve). (Right) Blasticidin (Bsd)-resistant CFU assay (1) using purified vpHAV containing a Bsd-selectable marker (vpHAV-Bsd) adsorbed to GL37 wt or GL37 HAVCR1 KO cells for 0 to 12 h. Briefly, monolayers were washed, incubated at 37°C for 24 h, trypsinized, and selected with Bsd. At 12 days postinfection, cell colonies were fixed and stained with crystal violet (dark spots), and 6-well plates were imaged using a flatbed scanner. (C) Quantitation of Bsd-resistant CFU in GL37 wt and GL37 HAVCR1 KO cells from panel B. Data are means ± SEM of colony numbers from four independent experiments. Differences between GL37 wt and GL37 HAVCR1 KO cells at each adsorption time point (black lines and asterisks) and between GL37 wt at different time points (blue lines and asterisks) were analyzed by Student’s unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Spread of HAV infection at different times postinfection in GL37 HAVCR1 KO cells transfected with HAVCR1 cDNA (KO-HAVCR1) or a vector (KO-vector). (Left) Expression of HAVCR1 at the cell surface of the transfectants by flow cytometry using anti-HAVCR1 MAb 1D12. The bar indicates HAVCR1-positive cells shown as a percentage. Transfectants were infected with HAV-Bsd at a multiplicity of infection (MOI) of 0.1 50% tissue culture infectious dose (TCID50)/cell, incubated at 37°C for 24 to 96 h, trypsinized, and plated in 6-well plates for a Bsd-resistant CFU assay (1). Results are representative of two independent experiments, each with two replicates. The number of Bsd-resistant CFU increased in GL37 wt cells and KO-HAVCR1 transfectants but not in KO-vector cells, indicating that the exo-HAV and vpHAV produced in infected cells cannot spread to other GL37 HAVCR1 KO cells in the absence of HAVCR1 expression. (E) Immunofluorescence analysis of HAV infection in HAVCR1 KO cells. GL37 wt cells and GL37 HAVCR1 KO cells transfected with a vector (KO-vector) or HAVR1 cDNA (KO-HAVCR1) were infected at a low MOI (0.1 TCID50/ml) of exo-HAV or vpHAV for 4 days, fixed, permeabilized, stained with anti-HAV neutralizing MAbs (green fluorescence), and counterstained with DAPI (4′,6-diamidino-2-phenylindole) nuclear dye (blue fluorescence). Micrographs of representative fields were taken at a magnification of ×400, showing that all GL37 wt cells are infected but that only ∼30% of HAVCR1 KO cells transfected with HAVCR1 cDNA were infected, which correlated with the number of transfected cells that express HAVCR1 at the cell surface (Fig. 1D, left) and indicated that exo-HAV and vpHAV cannot infect HAVCR1 KO cells in the absence of HAVCR1. (F) Gradient-purified exo-HAV not treated (green line) or treated with 1% Sarkosyl (red line) were repurified through iodixanol gradients. The HAV RNA in gradient fractions collected after isopycnic ultracentrifugation was quantified by reverse transcription-quantitative PCR (RT-qPCR), and the density of each fraction was determined by refractometry. Fractions 10 and 11, containing the peak of exo-HAV, were collected and used for further experimentation. Fraction 16, containing the peak of vpHAV extracted from exo-HAV by detergent treatment, was also collected. GE, genome equivalents. (G) Comparison of the infectivities in GL37 HAVCR1 KO cell transfectants of purified exo-HAV and naked HAV particles extracted from exo-HAV by detergent treatment. GL37 HAVCR1 KO cells transfected with HAVCR1 (KO-HAVCR1) or a vector (KO-vector) were infected with untreated exo-HAV (green bar), fractions 10 and 11 from panel F, or naked HAV particles extracted with detergent from exo-HAV (red bar), fraction 16 from panel F. Virus growth was determined by RT-qPCR at 72 h postinfection. Data are means ± SEM from three independent experiments. ns, nonsignificant.