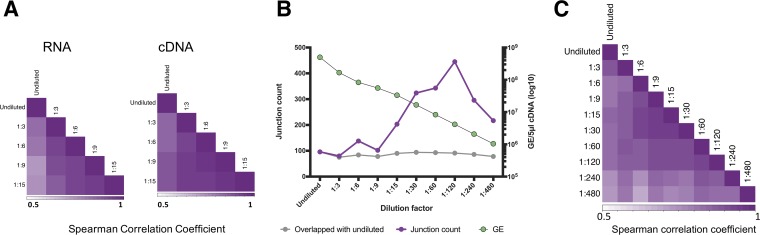
FIG 8.
Effects of viral template input on the detection of DIP-associated junctions. We serially diluted RNA or cDNA generated from the L1-P6-Rep1 sample and compared sequencing results between libraries generated with these dilutions as the templates. (A) Serial dilutions (1:3 to 1:15) was carried out on either the RNA or cDNA of the L1-P6-Rep1 sample, and a correlation test between the detected DIP-associated junctions was performed. (B) For each dilution, the total number of detected junctions (purple) is shown, along with the number of specific junctions that were also detected in the undiluted sample (gray). The copy number of viral cDNA molecules included in downstream PCR and library preparation for each dilution was determined by RT-qPCR (green; right y axis). GE, gene equivalent. (C) Read support values for all deletion junctions common across the diluted (at the cDNA level) and undiluted samples were normalized to the total number of deletion junction-spanning reads for each sample and used to perform a Spearman correlation between all pairs of samples using the R cor function.