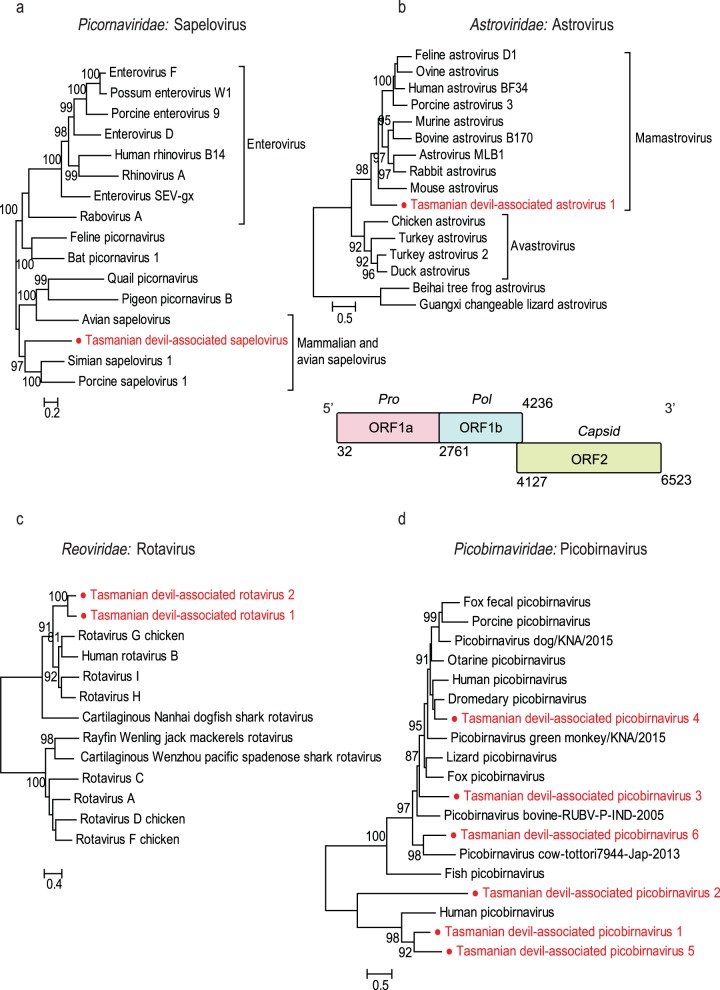
FIG 4.
Phylogenetic analyses and genomic structures of the RNA viruses identified in the feces of Tasmanian devils. All phylogenetic analyses were performed based on the amino acid sequence of the RdRp. (a) Tasmanian devil-associated sapelovirus; (b) Tasmanian devil-associated astrovirus; (c) Tasmanian devil-associated rotaviruses 1 and 2; (d) Tasmanian devil-associated picobirnaviruses 1 to 6. For Tasmanian devil-associated astrovirus, for which the whole-genome sequence was obtained, the genomic structure is shown below the corresponding phylogenetic tree. Predicted ORFs of these genomes are labeled with information of the potential protein or protein domain they encode. All trees are mid-point rooted and scaled to the number amino acid substitutions per site. Bootstrap values (>80%) are shown at the key nodes. The newly discovered viruses are shown in red in each phylogenetic tree.