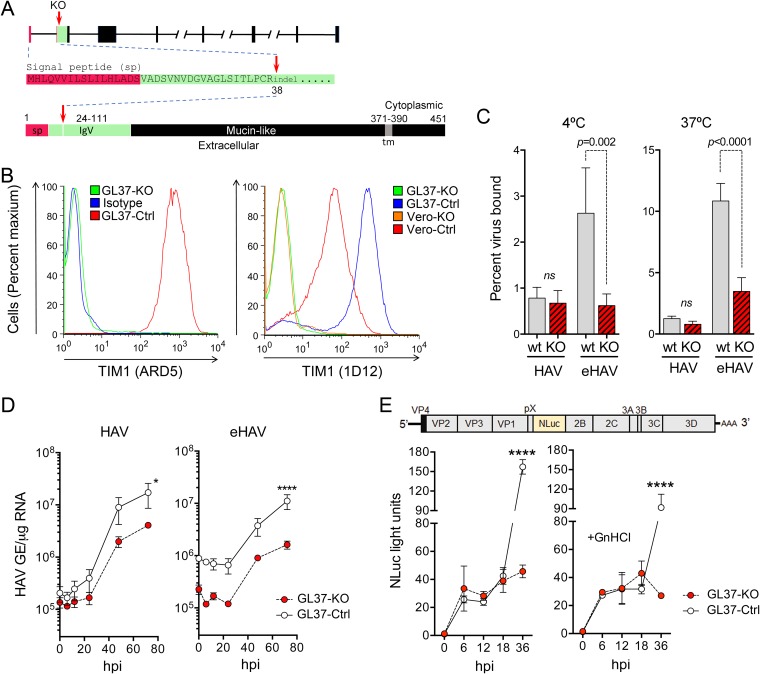
FIG 1.
GL37-KO cells lacking expression of TIM1 are permissive for HAV infection. (A) Organization of the TIM1 gene and its protein product in C. aethiops. Red arrows indicate where the sequence is disrupted (codon 38) with small indels in exon 2. At the bottom is shown the domain structure of TIM1 with amino acids numbered and the signal peptide (sp) sequence shown in red and the IgV domain (residues 24 to 111), which binds Ptd-Ser, in green. tm, transmembrane domain. (B) Flow cytometry analysis of cell surface staining for TIM1 with ARD5 (10) (left) and 1D12 (Biolegend) (right) monoclonal antibodies. Isotype, GL37-Ctrl cells stained with isotype control IgG. (C) Percent binding of iodixanol gradient-purified naked HAV and quasi-enveloped HAV to GL37-Ctrl and GL37-KO cells at 4°C (left) and 37°C (right). Results shown are means ± standard errors of the means (SEM) from 3 independent experiments, each with 2 to 3 technical replicates. ns, nonsignificant. (D) HAV RNA in lysates of GL37-Ctrl and GL37-KO cells following infection with gradient-purified naked (left) and quasi-enveloped (right) virus. GE, genome equivalents; hpi, hours postinoculation. Results are means of a total of 4 replicates ± SEM from 2 independent experiments. *, P = 0.017; ****, P < 0.0001 at 72 h. (E) At the top is shown the HM175/18f-NLuc genome organization, which has the nanoluciferase (NLuc) sequence inserted between the VP1pX and 2B coding sequences. Below is shown nanoluciferase activity in GL37-Ctrl and GL37-KO cell lysates following infection with detergent-treated HM175/18f-NLuc virus in the absence (left) or presence (right) of 5 mM guanidine hydrochloride (GnHCl). Nanoluciferase expression was insensitive to GnHCl between 6 and 18 hpi, indicating that it is due to translation of input naked viral genomes. Results are means of a total of 4 replicates ± SEM from 2 independent experiments. ****, P < 0.0001 at 72 h. All statistical testing was by two-way analysis of variance (ANOVA) with Sidak’s multiple-comparison test.