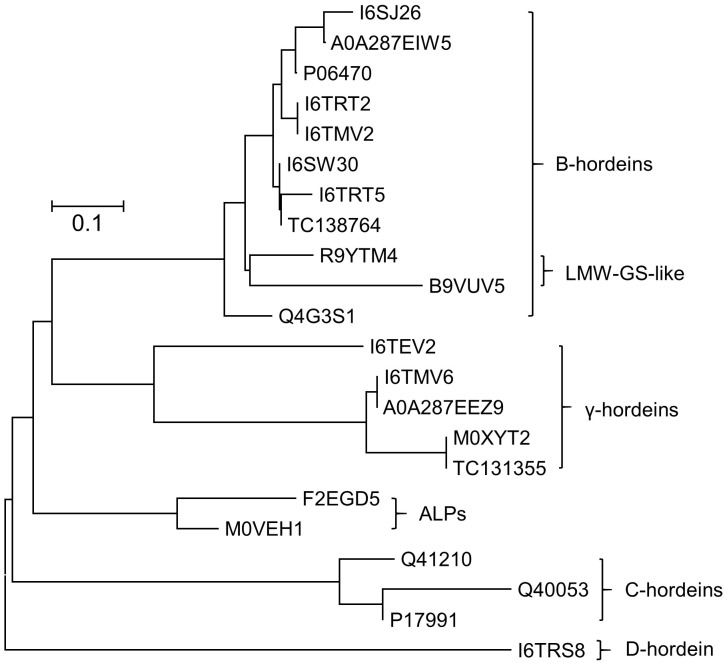
FIGURE 2.
Neighbor-joining phylogenetic tree showing the evolutionary relationship between hordein-like proteins present in H. vulgare. All sequences were retrieved from Uniprot database, except two proteins with a “TC” prefix which came from the TIGR database. Sequences were aligned using MUSCLE, and the analyzed in MEGAX (Kumar et al., 2018). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances that were used to infer the phylogenetic tree (scale bar, 0.1 amino acid substitutions per site). Two avenin-like proteins (ALPs) that share homology with the γ-hordeins were identified. Two novel B-hordein isoforms were identified based on peptides that map to wheat low molecular weight glutenins (LMW-GS), demonstrating that despite being absent in the barley genome, they were present in the barley lines studied.