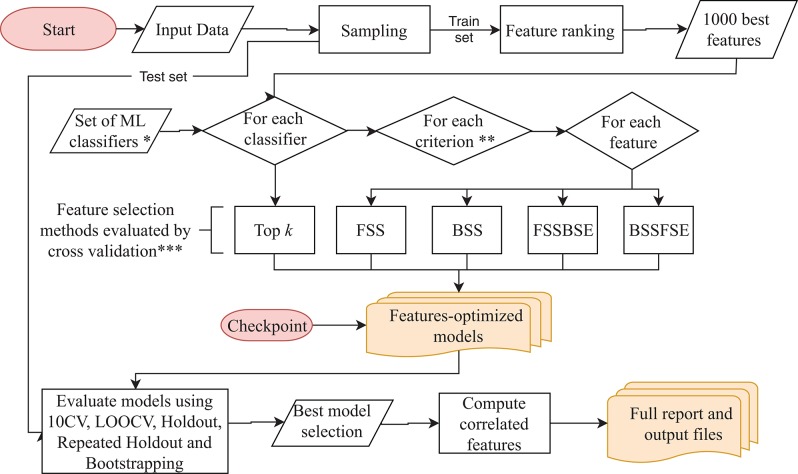
Figure 1.
BioDiscML pipeline. Preprocessing and feature selection procedures are fully parallelizable, When all features-optimized models are computed, the model selection starts. The program can be also started from the checkpoint at any moment during the execution. *The Set of ML classifiers is the set of pre-configured commands in classifiers.conf file. All classifiers are listed in the Supplementary Table S1. **Criterions are optimized metrics, evaluated by 10-folds cross validation (10 CV), used to assess if a model is improved, such as accuracy, balanced error rate, Matthew's correlation coefficient, area under the curve, sensitivity, specificity, Root Mean Squared Error, etc. (see Evaluation Criterion). ***Feature selection methods include forward stepwise selection (FSS), backward stepwise selection (BSS), forward stepwise selection and backward stepwise elimination (FSSBSE), backward stepwise selection, and Forward stepwise elimination (BSSFSE), and “top k” features (see Optimal Feature Subset Search Methods).