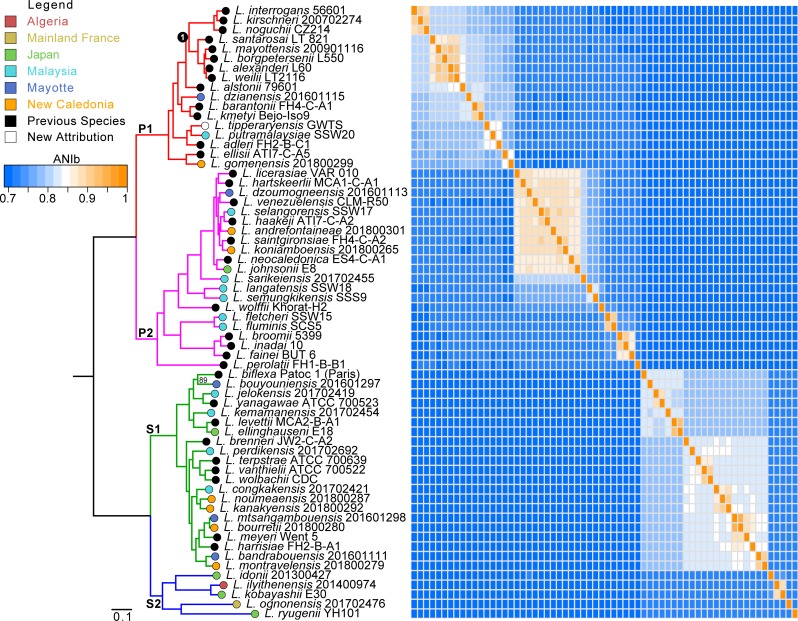
Fig 1. Phylogenetic tree based on the sequences of 1371 genes inferred as orthologous.
The matrix represents the calculated ANIb values for all the genomic sequences. The branches are colored according to their belonging to the four main subclades: P1 (red), P2 (purple), S1 (green) and S2 (blue). The bootstrap value is indicated for a single node (that corresponding to the separation between L. biflexa strain Patoc1 and L. bouyouniensis strain 201601297) since all the others have the maximum value of 100. A circle of color, according to the legend, represents the geographical origin of each of the new species described by this study. Node 1 indicates the node from which descent pathogenic species most frequently involved in human disease.