Abstract
C/D box snoRNAs (SNORDs) are a highly expressed class of non-coding RNAs. Besides their well-established role in rRNA modification, C/D box snoRNAs form protein complexes devoid of fibrillarin and regulate pre-mRNA splicing and polyadenylation of numerous genes. There is an emerging body of evidence for functional interactions between RNA viruses and C/D box snoRNAs. The infectivity of some RNA viruses depends on enzymatically active fibrillarin, and many RNA viral proteins associate with nucleolin or nucleophosmin, suggesting that viruses benefit from their cytosolic accumulation. These interactions are likely reflected by morphological changes in the nucleolus, often leading to relocalization of nucleolar proteins and ncRNAs to the cytosol that are a characteristic feature of viral infections. Knock-down studies have also shown that RNA viruses need specific C/D box snoRNAs for optimal replication, suggesting that RNA viruses benefit from gene expression programs regulated by SNORDs, or that viruses have evolved “new” uses for these humble ncRNAs to advance their prospects during infection.
1. snoRNAs are a highly expressed class of non-coding RNAs
Small nucleolar RNAs (snoRNAs) were first described in 1979 [1], making them one of the longest- and best-studied class of non-coding RNA (ncRNA). snoRNAs are highly expressed, for example SNORD3 expresses an estimated 200,000 copies/cell and SNORD13/14 express 20,000 molecules/cell [2,3], which compares to an estimated 200,000 total mRNA molecules per cell [4]. snoRNAs are 60–300 nt long. They accumulate in the nucleolus and are classified either as C/D box or H/ACA box snoRNAs. The best understood overall function of snoRNAs is to provide a scaffold that assembles a protein complex and to guide this complex to a target RNA, mainly rRNA, using base-pairing between the snoRNA and its target [[5], [6], [7], [8], [9]].
Recently, direct interactions between snoRNAs and pre-mRNAs as well as further processing of snoRNAs into miRNAs and shorter RNAs have emerged as new functions of snoRNAs [[10], [11], [12], [13]]. Here we review a new, so far understudied facet of snoRNA biology: the interaction of snoRNAs, mostly C/D box snoRNAs, with viruses.
C/D box snoRNAs (SNORDs) are typically 70–90 nt long and are characterized by several structural elements including the C (RUGAUGA) and D (CUGA) boxes, up to two antisense boxes that are complementary to the RNA target and complementary 5' and 3' ends that form a short terminal stem [14]. SNORDs form a protein complex consisting of NHP2L1 (aka 15.5k, SNU13), NOP56, NOP58 and fibrillarin [[15], [16], [17]] that catalyzes 2′-O-methylation of target rRNAs. The snoRNA acts as a scaffold for protein complex formation and also controls the recognition of other RNAs using antisense boxes that recognize sequences in ribosomal RNA resulting in 2′-O-methylation of the fifth nucleotide upstream of the D or D’ box by fibrillarin [14] (Fig. 1A and B).
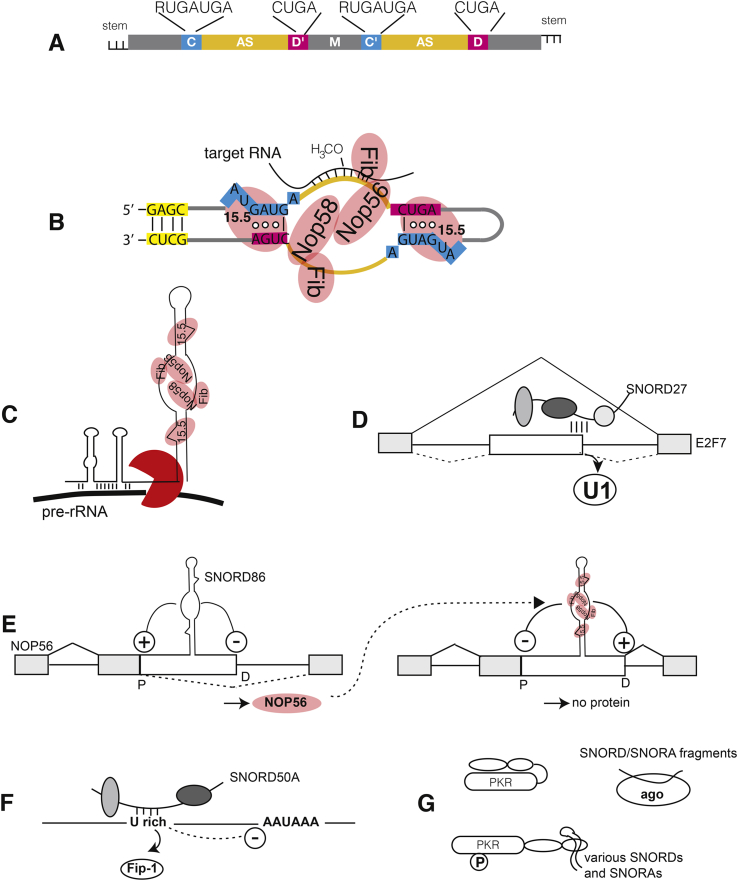
Fig. 1.
Molecular functions of snoRNAs.
A. Schematic structure of a C/D box snoRNAs. C, D and C′, D′ boxes are indicated with their consensus sequences, AS: antisense boxes, M: middle domain. In most cases SNORDs have short sequences exhibiting complementarity at the ends, which form short stems (see also B). B. Hypothetical structure of human snoRNP performing 2′-O-methylation. The SNORD forms a protein complex made of 15.5 (also known as SNU13 and NHP2L1), NOP56/58 and the methylase fibrillarin (Fib) that 2′-O-methylates (H3CO-) rRNA at a defined position (5 nt upstream of the D box). The coloring of the SNORD is similar to Fig. 1A. Circles indicate the base interaction within the RNA kink. Only one antisense box is shown in rRNA targeting, but both antisense boxes can be used. The structure is adopted from an archea snoRNP, based on NMR and cryo-EM studies [18,19]. C. SNORD3 guides endonucleases (red) to pre-rRNA, leading to cleavage. In addition to the C and D boxes, SNORD3 contains A and B boxes that interact with rRNA. D. Role of SNORDs in pre-mRNA splicing. SNORD27 binds to areas near an alternative 5′ splice site and blocks usage of this site through competition with U1 snRNP [20]. The constitutive exons are in gray, alternative exons are in white, the splicing patterns are indicated. E. Autoregulation of NOP56 formation due to alternative splicing. SNORD86 is located in an alternative 5′ exon (white box) of the NOP56 pre-mRNA. Without NOP56, the SNORD86 structure in the pre-mRNA activates a proximal splice site (P) and represses a distal (D) one, leading to exclusion of the alternative exon and the formation of a mRNA encoding NOP56 (left). The formation of a snoRNP containing NOP56 reverses this regulation, now blocking the proximal splice site and activating the distal one (right). The resulting mRNA does not encode a protein. An intermediate form that still contains R2TP proteins partially represses the proximal splice site (not shown) [21]. F. SNORD50A regulates polyadenylation. SNORD50A binds to the U-rich element that is part of the polyadenylation recognition site. SNORD50 binding removes FIP-1, a component of the cleavage and polyadenylation specificity factor (CPSF), which blocks polyadenylation at this site. Thus removal of SNORD50A increases usage of numerous alternative polyadenylation sites and increases expression of some mRNAs [22]. G. SNORDs bind to proteins. SNORDs and SNORAs bind to protein kinase RNA, leading to its activation, measured by PKR autophosphorylation (P) [23]. Fragments of SNORDs and SNORAs bind to Argonaute proteins, acting as miRNAs.
H/ACA box snoRNAs (SNORAs) have a conserved secondary structure composed of two hairpins, connected by a short hinge followed by a short tail. The hinge contains the conserved H box (‘hinge’) (ANANNA) and the tail harbors the ACA sequence [24]. SNORAs assemble a protein complex containing the pseudouridine synthetase Cbf5 (dyskerin) and the structural proteins NOP10, NHP2 and GAR1 [25]. SNORAs bind to rRNAs in the pseudouridylation pocket generated by the two hairpins, which allows recognition of the non-base paired substrate uridine that is isomerized to pseudouridine.
Ribosomal RNA (rRNA) is the main substrate of SNORDs and SNORAs. Around 100–106 2′-O-methylation [26,27] and around 100 pseudouridylation events are documented in rRNA [[28], [29], [30]]. These modifications are performed by SNORDs and SNORAs in the nucleolus.
In addition, the spliceosomal snRNAs U1, U2, U4 and U5 are modified by the related small Cajal body-specific RNA (scaRNA) in Cajal bodies [31]. Some scaRNAs are structurally indistinguishable from snoRNAs, whereas others combine C/D and H/ACA box motifs with other RNA parts [32].
The most abundant SNORD, SNORD3, contains an A and B box region upstream of the C and D boxes that bind to pre-rRNA. Despite the presence of fibrillarin in the SNORD3 complex, SNORD3 does not methylate pre-rRNA, but aids in its cleavage [33], (Fig. 1C).
2. Transcription, biogenesis and post-transcriptional modification of C/D box snoRNP particles
Recent analyses estimate the total number of human SNORDs to be between 357 and 417 [7]. The difference in the numbers is due to rules taking into account small RNA read coverage and mapping to the genome. Older databases, generated before deep sequencing data became available reported 264 snoRNAs [29]. Almost all of these SNORDs are intronic and localize in a hosting gene. However, a few SNORDs, notably SNORD3@ (@ indicating all four human SNORD3 genes), SNORD13, and SNORD118 (U8) are controlled by their own promoters [29].
Intron-encoded RNAs are released from pre-mRNA after the splicing reaction, where lariats are opened up by the debranching enzyme and are subsequently degraded through exonucleases [34,35]. SNORDs associate with proteins in a snoRNP precursor, where the SNORD's stem-termini form a short double strand structure that protects the RNA from endonucleases. The snoRNP assembly pathway has been unveiled for SNORD3 [36]. The R2TP complex (named after the yeast ATPases Rvb1 and Rvb2, Pih1 (Protein interacting with Hsp90) and Tah1 (TPR-containing protein associated with Hsp90) [37] acts like a chaperone and adds the proteins 15.5, NOP58, NOP56 and fibrillarin to the SNORDs in a stepwise fashion.
The most abundant snoRNA, SNORD3, is generated by its own pol II promoter and like all pol II transcripts, the primary SNORD3 RNA contains a N7-methylguanosine cap that is hypermethylated in the cytosol into a trimethyl guanosine capped RNA [38], which serves as a nuclear import signal. This demonstrates that some SNORDs are present at least temporarily in the cytosol. A cytosolic localization has been reported for numerous SNORDs under cellular stress conditions [39,40].
A subset of SNORDs undergoes post-transcriptional methylation at the N6 position of adenine residues located in the kink formed by C-D box interaction [41]. This modification prevents binding of the 15.5 protein to the kink and thus likely prevents the formation of a canonical snoRNP.
Biophysical studies showed that SNORD-RNPs exhibit a large structural diversity generated by forming dimers of a monomer SNORD-RNP (Fig. 1), which also suggests diverse functions [42].
3. Emerging novel molecular functions of C/D box snoRNAs
About half of the known SNORDs do not have predicted rRNA targets and are considered “orphan” [43,44], suggesting functions other than ncRNA-methylation or cleavage. Biochemical fractionation experiments suggest a structural diversity that indicates the molecular basis of novel functions. HeLa cell nuclei were separated under native salt conditions that leave native protein complexes intact. Some SNORDs copurified with spliceosomal fractions that are devoid of the methylase fibrillarin, indicating that 73 of 257 SNORDs tested act potentially outside the canonical rRNA methylation role [20]. Many of these SNORDs found in the fibrillarin-free fraction were known to act in rRNA methylation, indicating that SNORDs can have dual functions. The association of SNORDs with proteins other than fibrillarin, NOP56/58 and 15.5 has previously been demonstrated for SNORD115, which binds to hnRNPs [45] and can be biochemically separated in fractions containing and lacking fibrillarin [46]. Together, these data indicate that a given snoRNA can assemble into protein complexes containing a methylase (‘methylating complexes’) and into complexes lacking a methylase (‘non-methylating complexes’).
In both methylating and non-methylating complexes, the snoRNA will be protected from degradation by the associated proteins. The different complexes could also explain the occurrence of snoRNA fragments, which were first identified by RNAseq [47] and later also validated by RNase protection [45,48], as hnRNPs could cover a smaller region of the snoRNA than the NOP58/56/15.5 complex. These psnoRNAs (processed snoRNAs) are most prevalent for SNORD115 and SNORD116, which play a role in Prader-Willi syndrome [49], and the SNORD113, -114 families as well as SNORDs- 50, 19, 32B, 123, 111, 72 93, 23 and 85 [7,48]. Therefore, there are compelling reasons to suspect that SNORDs can fulfill new molecular functions different from rRNA modification.
Alternative pre-mRNA splicing has been identified as the first of these functions in SNORD115 (previously called HBII-52). SNORD115 is expressed in the Prader-Willi critical region and promotes inclusion of the alternative exon Vb of the serotonin receptor 2C exon due to direct SNORD:mRNA interaction [50]. Similarly, SNORD27 binds to the 5’ splice sites of the E2F7 transcripts and most likely blocks U1 snRNP access, leading to exon skipping [20] (Fig. 1D), and SNORD88C regulates the alternative splicing of the FGFR3 pre-mRNA [51]. In these examples, the SNORDs act in trans, i.e. they bind to a different pre-mRNA.
The NOP56 pre-mRNA is a fascinating example of a SNORD regulating alternative pre-mRNA splicing in cis. The NOP56 pre-mRNA hosts SNORD86 in an alternative 5’ exon (Fig. 1E). Within the pre-mRNA, SNORD86 forms a secondary structure that represses the distal splice site and activates the proximal splice site, resulting in exon skipping and formation of a NOP56 mRNA that encodes the NOP56 protein. Once enough NOP56 protein is formed, a snoRNP can form in the NOP56 pre-mRNA, which now activates the distal splice site and represses the proximal site, resulting in the inclusion of the alternative exon. Since this alternative exon causes a frameshift no functional NOP56 protein can be formed in the resulting RNA [21]. This system allows precise control of the formation of NOP56, which could be decisive for the formation of mature snoRNPs [36]. The regulation of the NOP56 alternative splicing through a secondary structure is reminiscent to the regulation of exon Vb/SNORD115 system, where the RNA structure dictates splicing [52].
Polyadenylation: SNORD50A influences polyadenylation by competing with the canonical FIP1 protein, which is a part of the cleavage and polyadenylation specificity factor (CPSF) that activates polyadenylation through binding to the U-rich sequence downstream of the polyadenylation site. Thus loss of SNORD50A promotes FIP1 binding leading to increased polyadenylation and up-regulation of gene expression and change in alternative polyadenylation site usage [22], (Fig. 1F).
Binding to proteins: Several SNORDs and SNORAs were found to bind and activate protein kinase R (PKR) [23]. PKR has been found to be activated by double-stranded RNAs that are generated in viral infections. Its activation generates an antiviral interferon response. Similarly, SNORD126 activates the PI3K-AKT pathway after activating FGFR2, but it is unclear whether this interaction is mediated by the snoRNA or proteins bound to the snoRNA [53]. SnoRNAs, both H/ACA box snoRNAs [54] and to a lesser extend C/D box snoRNAs [55,56] have been reported to be associated with Argonaute proteins, suggesting that fragments of a subset of snoRNAs can act as functional miRNAs. However, given the high expression of snoRNAs, the total number of snoRNAs cross-linked to Argonaute proteins was low, suggesting that miRNA precursors are minor function of snoRNAs [56], (Fig. 1G).
Targeting novel enzymatic activities: Unexpectedly, studies in yeast showed that two orphan SNORDs, snR4, snR45, guide acetylation of cytosine residues, rather than 2′-O-methylation to 18S rRNA, demonstrating that SNORDs can potentially guide other enzymes [57].
Effects of snoRNA on cellular processes. Loss or changes of SNORD expression has been associated with a large number of physiological changes. Notably SNORD expression is changed in cancer (reviewed in Ref. [58]), where in general SNORDs are upregulated and the types of SNORDs overexpressed correlated with cancer type and tumor stage [59]. As several of these SNORDs are orphans, their effects cannot be explained by changes in rRNA methylation.
Other physiological effects that cannot currently be explained by changes in rRNA methylation are downregulation of SNORD54 and 46 in lymphocytes of subjects with post traumatic stress disorder after the 9/11 attacks [60], changes of numerous SNORDs and SNORAs in circadian rhythm [61], and the regulation of systemic glucose metabolism by SNORDs that are hosted by the Rpl13a gene [62] (Table 1).
Table 1.
Changes of SNORD expression in diseases.
| SNORD54 and SNORD 46 | Downregulated in blood of persons with post traumatic stress disorder due to world trade center attacks | [60] |
| SNORD35b, 88, 57, 14d, SNORA17, 46, 17, 71 |
SNORDs cycle in circadian rhythm | [61] |
| SNORD32A, SNORD33, SNORD34, SNORD35A | SNORDs accumulate in the cytosol under lipotoxic stress and regulate systemic glucose metabolism | [62] |
| SNORD126 | Increases fibroblast growth factor receptor 2 (FGFR2), leading increased phosphorylation of AKT, GSK-3beta, p70S6K | [53] |
| SNORD50A | Blocks FIP1 binding to the downstream U-rich sequence of the polyadenylation site, loss of SNORD50A leads to increased polyadenylation and up-regulation of gene expression | [22] |
| SNORD115 and SNORD116 clusters | Missing in Prader-Willi syndrome | [49] |
| Numerous SNORDs | Deregulated in cancer | [59] |
Reflecting the broader functions of snoRNAs, it is not surprising that viruses, especially RNA viruses use snoRNAs for their purposes. We summarize here the evidence that accumulated over the last decade describing an interplay between SNORDs and viruses.
4. Viruses use SNORDs for infectivity
Experiments using gene-trap insertional mutagenesis provided the first evidence for the role of SNORDs in viral infections. Twelve different viruses were tested in various cell lines expressing gene trap libraries that knocked down individual host genes. This screen identified a total of 83 SNORDs and SNORAs as factors necessary for viral infectivity. The eight SNORDs with the strongest effect across all viruses are hosted by the non-protein coding SNHG1 transcript. Three of the SNORDs are expressed by their own promoter, indicating that the SNORDs, not the hosting genes were necessary for the replication of several DNA viruses (CPV, HSV2) and RNA viruses (DFV, FLU, HRV16, and RSV) [63]. This interplay between viruses and SNORDs is found in numerous individually studied viruses. For example, SNORD3@, SNORD44, SNORD76 and SNORD78 (previously called U3, U44, U76, U78) are upregulated after viral infection by Chikungunya fever virus (CHIKV) [64]; and significant SNORD expression changes were observed in pig blood after infection with porcine reproductive and respiratory syndrome virus (PRRSV) [65].
Among the herpesviruses, which have relatively large DNA genomes, there are several published accounts of SNORD expression or processing changes upon infection. SNORD-fragments that potentially act as miRNAs appear in cells after infection with bovine herpesvirus 1 [66]; and infection with another herpesvirus, murine cytomegalovirus, downregulates numerous SNORDs [67].
Most viruses do not encode their own snoRNAs. The exception is the Epstein-Barr herpesvirus (EBV), a DNA virus causing mononucleosis. EBV encodes a viral-specific v-snoRNA1 that is processed into smaller RNAs, which could have miRNA-like functions. However, despite the strong upregulation of v-snoRNA1 during infection, its deletion had no obvious phenotype, so its function remains unclear [68].
Finally, there are numerous accounts of retroviruses incorporating non-coding RNAs, including SNORDs into their virions. For example, murine leukemia virus packages host non-coding RNAs and selectively incorporates SNORD104 into its virions [67]. Likewise, the packageome of Maloney leukemia virus [69] and human immunodeficiency virus type 1 [70] contains overrepresented SNORDs and other ncRNAs. It has been hypothesized that these ncRNAs may help packaging of the virions [69], but detailed functions remain to be determined.
With the exceptions of MCMV (murine cytomegalovirus) and EBV, the reports describing an interaction between SNORDs and viruses are limited to RNA viruses, suggesting that mainly RNA viruses use SNORDs. Despite these reports, very little is known about how a virus benefits from SNORD interactions on a molecular basis. Direct interactions of viral proteins with fibrillarin, the RNA methylase associated with SNORDs have been reported. In addition RNA viruses add the methyl-guanosine caps of several SNORDs to their own RNAs using cap-snatching.
5. Direct interactions of viruses with snoRNAs and their associated proteins
snoRNAs are mostly localized in the nucleolus, although a subset of snoRNAs can localize to the cytosol as a cellular response to specific environmental stimuli [40] and SNORDs transcribed by pol II, like SNORD3, are exported into the cytosol where their cap is hypermethylated, leading to their nuclear re-import.
Most RNA viruses replicate in the cytoplasm. They bring with them (negative sense viruses) or encode (positive sense viruses) an RNA dependent RNA polymerase that carries out replication and transcription activities in the cytosol independent of the host cell polymerases, which are located in the nucleus. A few RNA viruses replicate within the nucleus, and therefore their RNA-dependent RNA polymerases are nuclear. Viral mRNAs are translated by host ribosomes either in the cytosol, in localized virus factories often associated with the Golgi apparatus, and/or on the rough endoplasmic reticulum (Fig. 2). Despite the diversity of replication sites and strategies among different viral species, there is evidence for direct interactions between snoRNAs and/or their associated proteins with viruses.
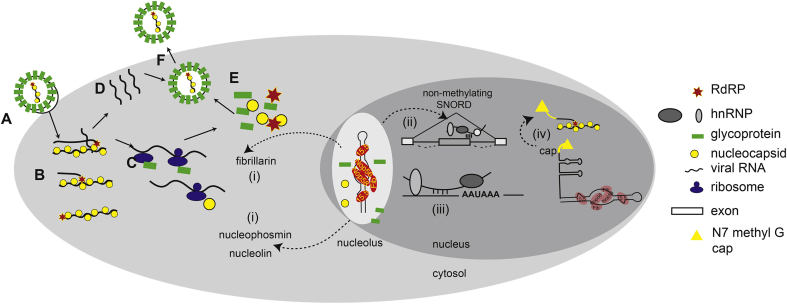
Fig. 2.
Replication cycle of a generic RNA virus.
Although specific steps of viral replication cycle may differ between virus families, the requisite discrete steps are outlined here. A. Virus attaches to the target host cell via interactions of surface exposed glycoproteins (green bars) and host cell surface proteins that serve as viral receptors. B. The virus particle is internalized via endocytosis or the viral genome is released into the cytoplasm via fusion of the viral and cellular membranes. Inside the cell the viral genome, typically coated with a viral nucleocapsid protein (yellow balls), the genomic RNA is transcribed by the viral RNA-dependent RNA polymerase (RDRP; red stars) to make mRNA (for negative sense viruses), or the genome serves directly as mRNA (for positive sense viruses). C. The mRNAs are translated by host cell ribosomes, either in the cytosol, within concentrated areas of viral production (so called virus factories, often associated with the Golgi) or on the rough ER (e.g., for glycoproteins that require further processing). D. The viral genome is replicated with the virally encoded RDRP (RNA dependent RNA polymerase). E. Newly synthesized viral proteins and viral genomes self-assemble into new virus particles. F. New virions are released from the cell via exocytosis, fusion with the cell membrane, or upon cell lysis.Viral components may access host snoRNAs either when the snoRNAs are present in the cytosol, when viral proteins possess nuclear or nucleolar localization signals, or in the special cases when the virus employs a replication strategy that included nuclear localization, e.g., the orthomyxoviruses, bornaviruses, or retroviruses. During infection, viruses can cause components of the SNORD methylating complex to be re-purposed. For example, i. viral infection of some viruses cause translocation of fibrillarin, nucleophosmin or nucleolin from the nucleolus to the cytosol. ii. SNORDs normally employed for rRNA modification in methylation complexes in the nucleolus can be re-trafficked into the nucleoplasm where they influence splice site selection or, iii., polyadenylation and mRNA stability; iv. Nascent transcripts of SNORDs that are independently transcribed can be exploited by viruses for cap-snatching, transferring a N7methyl guanosine cap to the 5′ end of a viral RNA.
5.1. Interaction with fibrillarin
Fibrillarin is an RNA methyltransferase associated with all methylating SNORDs (Fig. 1B). The protein is concentrated in the dense fibrillar component of the nucleolus. Fibrillarin is evolutionary highly conserved and contains three domains: an N-terminal glycine-arginine-rich domain (GAR) interacting with numerous proteins, a central domain that binds RNA and a C-terminal domain that binds to NOP56. The methyltransferase activity is localized to the central and C-terminal domain [71].
Porcine reproductive and respiratory syndrome virus (PRRSV) is an RNA virus that replicates in the cytosol. However, its nucleocapsid protein (N) accumulates in the nucleus and nucleolus where N binds to the GAR-domain of fibrillarin in an RNA-dependent manner [72]. Similarly, the ORF3 protein from the plant umbravirus and groundnut rosette virus forms a stoichiometric 1:1 complex with fibrillarin. ORF3 interacts with fibrillarin in the nucleolus and then causes the relocalization of fibrillarin into the cytosol, eventually allowing the virus to spread through the entire plant through the plant phloem using ORF3-fibrillarin RNP complexes [73].
A genome-wide screen for host genes required for heniapavirus infectivity identified fibrillarin as a host factor that is important for infection. Rescue experiments showed that the methylase activity of fibrillarin was necessary for infectivity. Several related viruses that replicate in the cytosol (measles, mumps and respiratory syncytial virus) showed a similar dependency on fibrillarin, whereas influenza viruses that replicate predominantly in the nucleus were less affected [74]. Fibrillarin is not required for virus entry, but is important for the expression of two abundant viral proteins, matrix (M), and nucleocapsid, (N), suggesting that fibrillarin is necessary for the translation of viral proteins.
5.2. Viruses snatch caps from SNORDs
Cellular mRNAs harbor a N7-methylguanylate cap (m7G) that is necessary for translation, RNA processing and export. In general, DNA viruses use the cellular capping machinery for viral RNAs. Most RNA viruses cannot use this mechanism as they replicate in the cytosol without access to the cellular capping machinery. Thus RNA viruses have developed different strategies to overcome their capping problem.
Predominantly plant viruses (carmoviruses, tombusviruses) can translate from RNAs that have triphosphate 5’ termini. Picornaviruses and birnaviruses covalently link their mRNAs to small proteins to initiate translation [75]. Many other positive and negative sense RNA viruses possess capping activity within their RNA dependent RNA polymerases or express a separately encoded enzyme [76,77].
Another viral strategy to overcome the capping problem is called ‘cap-snatching’, which entails the use of a short capped host RNA fragment to prime synthesis of viral RNA. Cap-snatching was first discovered in influenza virus [78] and has since been described for both nuclear and cytosolic viruses. Global RNA sequencing of human epithelial cells infected with influenza A virus (IAV H1N1/swine flu) [79] and IAV (Brisbane/59/2007) [80] showed that the viruses snatch caps from numerous ncRNAs but also from snoRNAs that are controlled by their own promoter: SNORD3, SNORD118 (U8) and SNORD13. As pol II transcripts, these SNORDs contain a m7G cap in their primary transcript that is exported into the cytosol where the cap is hypermethylated into a 2,2,7-trimethyl cap leading to nuclear re-import. Interestingly, the viral caps were m7G caps that support translation. Since the influenza polymerase interacts with the polymerase pol II carboxy terminal domain (CTD) in the nucleus [81], it is likely that the primary snoRNAs are snatched in the nucleus before their cytosolic export.
6. Many viruses access the nuclear/nucleolar compartments
Although most RNA viruses replicate in the cytoplasm, some replicate within the nucleus. Nevertheless, many viruses seek access to the nucleus and the nucleolus with nuclear localization signals or simple diffusion of small viral proteins through the nuclear pores. The interaction factors for many viral proteins are host factors are normally associated with snoRNA function or processing.
A few animal RNA virus families, namely the Orthomyxoviridae (e.g., influenza virus), Nyamiviridae (e.g., Midway virus), Bornaviridae (e.g. borna disease virus), and Retroviridae (e.g., HIV-1), replicate entirely or employ some replicative steps within the nuclei of their host cells. This nuclear lifestyle offers additional opportunities for viral components to interact with snoRNAs that are abundant in the nucleoplasm or the nucleoli.
Many animal/human RNA viruses encode proteins that possess a nuclear or nucleolar localization signal sequence [[82], [83], [84], [85], [86], [87]]. In particular, there are abundant published accounts of viral nucleocapsid proteins that localize at least transiently to the nucleus (reviewed recently in Ref. [88]). Nuclear or nucleolar localization affords access to a much richer supply of snoRNAs for these viral proteins. Furthermore, many proteins encoded by RNA viruses are small enough to enter the nucleus. It is known that proteins smaller than ∼50 kDa can diffuse freely into the nucleus, even without a localization signal [89] and thus viral proteins can interact at least transiently with the nucleolar SNORDs.
Viruses and nucleolar proteins: The interactions of viruses and their proteins with the host cell nucleolus has been studied extensively (see Refs. [[90], [91], [92], [93]] for excellent reviews on this broad topic). The reason why proteins encoded by the mostly cytoplasmic-replicating viruses would localize to the nucleolus is unclear. One possibility is that the virus co-opts components of the nucleolus for use in its replication strategy, or that interference with a normal nucleolar function improves viral replication. In addition to the role of fibrillarin in viral infectivity described earlier, the interaction with two predominant nucleolar proteins, nucleophosmin and nucleolin, are best documented.
Nucleophosmin, (NPM1, also called B23) functions like a chaperone mediating interactions between RNA and proteins. NPM1 has many demonstrated binding partners, including RNA, DNA, nucleolar proteins nucleolin, p53, and transcription factors, but its ability to shuttle between the cytoplasm, nucleoplasm, and nucleolus may be the defining feature that makes it an attractive target for viral proteins [94] and NPM1 has been implicated in several distinct interactions with different viruses.
For example, in bovine immunodeficiency virus, the Rev protein has nuclear and nucleolar localization signals. Rev requisitely interacts with nucleophosmin and its knockdown inhibits viral replication [95]. This recent observation mirrors what was seen previously in HIV-1, where Rev was shown to interact with NPM1 and localize to the nucleolus [96]. Porcine epidemic diarrhea virus (PEDV, a positive sense RNA virus) nucleocapsid protein N localizes to nucleolus and interacts with NPM1. It is thought that NPM1 protects the nucleocapsid protein N from proteolytic cleavage independent of NPM1's nucleolar regular functions [97]. The replication of nervous necrosis virus (nodavirus; a positive sense RNA virus) was also highly dependent on NPM1, which appeared to help shuttle the viral capsid protein across the nuclear membrane [98]. Localization of the matrix protein of the paramyxoviruses (negative sense RNA viruses) to the nucleolus appears important for viral replication and this translocation is facilitated by interaction with NPM1 during Newcastle disease virus infection [99].
Nucleolin. For viral-nucleolar protein interactions, it is notable that the largest number of published observations relate to nucleolin (NCL) (reviewed recently in Ref. [92]). These interactions have been observed at almost every step of viral replication cycles, including binding and entry, genome replication, transcription, and viral assembly, transport, and egress. The diversity of functions for which viruses exploit this protein is probably due to nucleolin's ability to localize on the cell surface, as well as within the cytoplasm, nucleoplasm, and nucleolus.
The naturally abundant expression of nucleolin and nucleophosmin could lead to interactions with viral proteins that are not physiologically relevant. However, the repeated discovery of viral protein interaction with nucleolar proteins in a wide diversity of systems suggests that at least some of these interactions are important for virus propagation, the host antiviral response, or both.
7. Hypothesis: RNA viruses repurpose nucleolar proteins and RNAs
The evidence reviewed here supports an emerging model where RNA viruses repurpose nucleolar proteins for their propagation. A common feature of RNA virus infection is a morphological change to the nucleolus, resulting in the accumulation of nucleophosmin, nucleolin and fibrillarin in the cytosol. Nucleophosmin is an acidic protein that acts like a chaperone mediating interactions between RNA and proteins [94]. Similarly, nucleolin is a multifunctional RNA binding protein with an intrinsic DNA and RNA helicase activity involved in arranging RNA:protein complexes [100]. RNA viruses form cytosolic replication centers and inclusion bodies and it is possible that nucleophosmin and nucleolin organize the assembly of viral RNA protein complexes, functions that these proteins have in rRNA biosynthesis. Such an organizing function has been shown for CMV replication compartments [101]. In addition, infection of some viruses relocalize fibrillarin from the nucleus to the cytosol, where host fibrillarin could act on yet unidentified RNAs [73].
Many SNORDs can form complexes containing or lacking fibrillarin, and it is possible that when viral proteins disrupt methylating complexes and as fibrillarin is relocalized and accumulates in the cytosol, the relative concentration of fibrillarin-free (‘non-methylating’) complexes is increased. Non-methylating SNORDs act on pre-mRNA splicing and polyadenylation (Fig. 1). Numerous studies showed a selective change in SNORD expression after viral infection, and it is thus possible that these SNORDs change host or viral mRNA splicing patterns and/or polyadenylation site usage in a way that is beneficial for the virus. Finally, a change in SNORD-RNP complexes could affect the ability of snoRNAs to activate protein kinase R, which could modulate the host immune response [9,23].
The examples highlighted here demonstrate that co-evolution of viral parasites with their host cells has resulted in a diversity of creative ways by which viruses have co-opted SNORDs to enhance every step of viral propagation from entry into the cell through evasion of host response to egress. Studying the association between SNORDs and viruses will likely uncover new functions of SNORDs possibly unveil new anti-viral strategies.
Acknowledgements
We would like to gratefully acknowledge funding for this research by NIH grants NIGMS P20GM103546 and AI137620 (JSL) grant number R21NS09818601-A1 (SS) and a grant by the Foundation for Prader-Willi Research to SS.
Contributor Information
Stefan Stamm, Email: stefan@stamms-lab.net.
J. Stephen Lodmell, Email: stephen.lodmell@umontana.edu.
References
- 1.Reddy R., Henning D., Busch H. Nucleotide sequence of nucleolar U3B RNA. J. Biol. Chem. 1979;254:11097–11105. [PubMed] [Google Scholar]
- 2.Maxwell E.S., Fournier M.J. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 3.Tyc K., Steitz J.A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapiro E., Biezuner T., Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 5.Bratkovic T., Rogelj B. The many faces of small nucleolar RNAs. Biochim. Biophys. Acta. 2014;1839:438–443. doi: 10.1016/j.bbagrm.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Dupuis-Sandoval F., Poirier M., Scott M.S. vol. 6. Wiley Interdiscip Rev RNA; 2015. pp. 381–397. (The Emerging Landscape of Small Nucleolar RNAs in Cell Biology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jorjani H., Kehr S., Jedlinski D.J., Gumienny R., Hertel J., Stadler P.F., Zavolan M., Gruber A.R. An updated human snoRNAome. Nucleic Acids Res. 2016;44:5068–5082. doi: 10.1093/nar/gkw386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massenet S., Bertrand E., Verheggen C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2016:1–13. doi: 10.1080/15476286.2016.1243646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stepanov G.A., Filippova J.A., Nushtaeva A.A., Kuligina E.V., Koval O.A., Richter V.A., Semenov D.V. Artificial analogues of circulating box C/D RNAs induce strong innate immune response and MicroRNA activation in human adenocarcinoma cells. Adv. Exp. Med. Biol. 2016;924:121–125. doi: 10.1007/978-3-319-42044-8_24. [DOI] [PubMed] [Google Scholar]
- 10.Dupuis-Sandoval F., Poirier M., Scott M.S. vol. 6. Wiley Interdiscip Rev RNA; 2015. pp. 381–397. (The Emerging Landscape of Small Nucleolar RNAs in Cell Biology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falaleeva M., Welden J.R., Duncan M.C., Stamm S. C/D-box snoRNAs form methylating and non methylating ribonucleoprotein complexes: old dogs show new tricks. Bioessays. 2017;39 doi: 10.1002/bies.201600264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mleczko A.M., Bakowska-Zywicka K. When small RNAs become smaller: emerging functions of snoRNAs and their derivatives. Acta Biochim. Pol. 2016;63:601–607. doi: 10.18388/abp.2016_1330. [DOI] [PubMed] [Google Scholar]
- 13.Shi J., Huang C., Huang S., Yao C. snoRNAs associate with mRNA 3' processing complex: new wine in old bottles. RNA Biol. 2018;15:194–197. doi: 10.1080/15476286.2017.1416278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filipowicz W., Pelczar P., Pogacic V., Dragon F. Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochim. Pol. 1999;46:377–389. V. [PubMed] [Google Scholar]
- 15.Filipowicz W., Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 16.Hirose T., Shu M.D., Steitz J.A. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell. 2003;12:113–123. doi: 10.1016/s1097-2765(03)00267-3. [DOI] [PubMed] [Google Scholar]
- 17.Watkins N.J., Bohnsack M.T. vol. 3. Wiley Interdiscip Rev RNA; 2012. pp. 397–414. (The Box C/D and H/ACA snoRNPs: Key Players in the Modification, Processing and the Dynamic Folding of Ribosomal RNA). [DOI] [PubMed] [Google Scholar]
- 18.Lapinaite A., Simon B., Skjaerven L., Rakwalska-Bange M., Gabel F., Carlomagno T. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature. 2013;502:519–523. doi: 10.1038/nature12581. [DOI] [PubMed] [Google Scholar]
- 19.Yip W.S., Shigematsu H., Taylor D.W., Baserga S.J. Box C/D sRNA stem ends act as stabilizing anchors for box C/D di-sRNPs. Nucleic Acids Res. 2016;44:8976–8989. doi: 10.1093/nar/gkw576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falaleeva M., Pages A., Matuszek Z., Hidmi S., Agranat-Tamir L., Korotkov K., Nevo Y., Eyras E., Sperling R., Stamm S. Dual function of C/D box snoRNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1625–E1634. doi: 10.1073/pnas.1519292113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lykke-Andersen S., Ardal B.K., Hollensen A.K., Damgaard C.K., Jensen T.H. Box C/D snoRNP Autoregulation by a cis-Acting snoRNA in the NOP56 Pre-mRNA. Mol. Cell. 2018;72:99–111. doi: 10.1016/j.molcel.2018.08.017. e115. [DOI] [PubMed] [Google Scholar]
- 22.Huang C., Shi J., Guo Y., Huang W., Huang S., Ming S., Wu X., Zhang R., Ding J., Zhao W. A snoRNA modulates mRNA 3' end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Res. 2017;45:8647–8660. doi: 10.1093/nar/gkx651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Youssef O.A., Safran S.A., Nakamura T., Nix D.A., Hotamisligil G.S., Bass B.L. Potential role for snoRNAs in PKR activation during metabolic stress. Proc. Natl. Acad. Sci. U. S. A. 2015;112:5023–5028. doi: 10.1073/pnas.1424044112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganot P., Caizergues-Ferrer M., Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 25.Lafontaine D.L., Bousquet-Antonelli C., Henry Y., Caizergues-Ferrer M., Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogh N., Jansson M.D., Hafner S.J., Tehler D., Birkedal U., Christensen-Dalsgaard M., Lund A.H., Nielsen H. Profiling of 2'-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 2016;44:7884–7895. doi: 10.1093/nar/gkw482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y., Pirnie S.P., Carmichael G.G. High-throughput and site-specific identification of 2'-O-methylation sites using ribose oxidation sequencing (RibOxi-seq) RNA. 2017;23:1303–1314. doi: 10.1261/rna.061549.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew Chem. Int. Ed. Engl. 2015;54:451–455. doi: 10.1002/anie.201408362. [DOI] [PubMed] [Google Scholar]
- 29.Lestrade L., Weber M.J. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piekna-Przybylska D., Decatur W.A., Fournier M.J. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darzacq X., Jady B.E., Verheggen C., Kiss A.M., Bertrand E., Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2'-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tycowski K.T., Aab A., Steitz J.A. Guide RNAs with 5' caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr. Biol. 2004;14:1985–1995. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Granneman S., Vogelzangs J., Luhrmann R., van Venrooij W.J., Pruijn G.J., Watkins N.J. Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol. Cell Biol. 2004;24:8600–8610. doi: 10.1128/MCB.24.19.8600-8610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costello J.L., Stead J.A., Feigenbutz M., Jones R.M., Mitchell P. The C-terminal region of the exosome-associated protein Rrp47 is specifically required for box C/D small nucleolar RNA 3'-maturation. J. Biol. Chem. 2011;286:4535–4543. doi: 10.1074/jbc.M110.162826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miki T.S., Grosshans H. The multifunctional RNase XRN2. Biochem. Soc. Trans. 2013;41:825–830. doi: 10.1042/BST20130001. [DOI] [PubMed] [Google Scholar]
- 36.Bizarro J., Charron C., Boulon S., Westman B., Pradet-Balade B., Vandermoere F., Chagot M.E., Hallais M., Ahmad Y., Leonhardt H. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J. Cell Biol. 2014;207:463–480. doi: 10.1083/jcb.201404160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kakihara Y., Saeki M. The R2TP chaperone complex: its involvement in snoRNP assembly and tumorigenesis. Biomol. Concepts. 2014;5:513–520. doi: 10.1515/bmc-2014-0028. [DOI] [PubMed] [Google Scholar]
- 38.Lubben B., Marshallsay C., Rottmann N., Luhrmann R. Isolation of U3 snoRNP from CHO cells: a novel 55 kDa protein binds to the central part of U3 snoRNA. Nucleic Acids Res. 1993;21:5377–5385. doi: 10.1093/nar/21.23.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brandis K.A., Gale S., Jinn S., Langmade S.J., Dudely-Rucker N., Jiang H., Sidhu R., Ren A., Goldberg A., Schaffer J.E. Box C/D snoRNA U60 regulates intracellular cholesterol trafficking. J. Biol. Chem. 2013;288:35703–35713. doi: 10.1074/jbc.M113.488577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holley C.L., Li M.W., Scruggs B.S., Matkovich S.J., Ory D.S., Schaffer J.E. Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase. J. Biol. Chem. 2015;290:11741–11748. doi: 10.1074/jbc.M115.637413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang L., Ashraf S., Wang J., Lilley D.M. Control of box C/D snoRNP assembly by N(6)-methylation of adenine. EMBO Rep. 2017;18:1631–1645. doi: 10.15252/embr.201743967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G., Zhao Y., Li H. The multistructural forms of box C/D ribonucleoprotein particles. RNA. 2018;24:1625–1633. doi: 10.1261/rna.068312.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deschamps-Francoeur G., Garneau D., Dupuis-Sandoval F., Roy A., Frappier M., Catala M., Couture S., Barbe-Marcoux M., Abou-Elela S., Scott M.S. Identification of discrete classes of small nucleolar RNA featuring different ends and RNA binding protein dependency. Nucleic Acids Res. 2014;42:10073–10085. doi: 10.1093/nar/gku664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith C.M., Steitz J.A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 45.Kishore S., Khanna A., Zhang Z., Hui J., Balwierz P.J., Stefan M., Beach C., Nicholls R.D., Zavolan M., Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010;19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soeno Y., Taya Y., Stasyk T., Huber L.A., Aoba T., Huttenhofer A. Identification of novel ribonucleo-protein complexes from the brain-specific snoRNA MBII-52. Rna. 2010;16:1293–1300. doi: 10.1261/rna.2109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taft R.J., Glazov E.A., Lassmann T., Hayashizaki Y., Carninci P., Mattick J.S. Small RNAs derived from snoRNAs. Rna. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen M., Eyras E., Wu J., Khanna A., Josiah S., Rederstorff M., Zhang M.Q., Stamm S. Direct cloning of double-stranded RNAs from RNase protection analysis reveals processing patterns of C/D box snoRNAs and provides evidence for widespread antisense transcript expression. Nucleic Acids Res. 2011;39:9720–9730. doi: 10.1093/nar/gkr684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cavaille J. Wiley Interdiscip Rev RNA; 2017. Box C/D Small Nucleolar RNA Genes and the Prader-Willi Syndrome: a Complex Interplay. [DOI] [PubMed] [Google Scholar]
- 50.Kishore S., Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 51.Scott M.S., Ono M., Yamada K., Endo A., Barton G.J., Lamond A.I. Human box C/D snoRNA processing conservation across multiple cell types. Nucleic Acids Res. 2012;40:3676–3688. doi: 10.1093/nar/gkr1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen M., Bellaousov S., Hiller M., de La Grange P., Creamer T.P., Malina O., Sperling R., Mathews D.H., Stoilov P., Stamm S. Pyrvinium pamoate changes alternative splicing of the serotonin receptor 2C by influencing its RNA structure. Nucleic Acids Res. 2013;41:3819–3832. doi: 10.1093/nar/gkt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang X., Yang D., Luo H., Wu S., Dong W., Xiao J., Yuan S., Ni A., Zhang K.J., Liu X.Y. SNORD126 promotes HCC and CRC cell growth by activating the PI3K-AKT pathway through FGFR2. J. Mol. Cell Biol. 2017;9:243–255. doi: 10.1093/jmcb/mjw048. [DOI] [PubMed] [Google Scholar]
- 54.Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 55.Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 56.Kishore S., Gruber A.R., Jedlinski D.J., Syed A.P., Jorjani H., Zavolan M. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol. 2013;14:R45. doi: 10.1186/gb-2013-14-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma S., Yang J., van Nues R., Watzinger P., Kotter P., Lafontaine D.L.J., Granneman S., Entian K.D. Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mannoor K., Liao J., Jiang F. Small nucleolar RNAs in cancer. Biochim. Biophys. Acta. 2012;1826:121–128. doi: 10.1016/j.bbcan.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong J., Li Y., Liu C.J., Xiang Y., Li C., Ye Y., Zhang Z., Hawke D.H., Park P.K., Diao L. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep. 2017;21:1968–1981. doi: 10.1016/j.celrep.2017.10.070. [DOI] [PubMed] [Google Scholar]
- 60.Kuan P.F., Waszczuk M.A., Kotov R., Clouston S., Yang X., Singh P.K., Glenn S.T., Cortes Gomez E., Wang J., Bromet E. Gene expression associated with PTSD in World Trade Center responders: an RNA sequencing study. Transl. Psychiatry. 2017;7:1297. doi: 10.1038/s41398-017-0050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aitken S., Semple C.A. The circadian dynamics of small nucleolar RNA in the mouse liver. J. R. Soc. Interface. 2017;14 doi: 10.1098/rsif.2017.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee J., Harris A.N., Holley C.L., Mahadevan J., Pyles K.D., Lavagnino Z., Scherrer D.E., Fujiwara H., Sidhu R., Zhang J. Rpl13a small nucleolar RNAs regulate systemic glucose metabolism. J. Clin. Invest. 2016;126:4616–4625. doi: 10.1172/JCI88069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray J.L., Sheng J., Rubin D.H. A role for H/ACA and C/D small nucleolar RNAs in viral replication. Mol. Biotechnol. 2014;56:429–437. doi: 10.1007/s12033-013-9730-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saxena T., Tandon B., Sharma S., Chameettachal S., Ray P., Ray A.R., Kulshreshtha R. Combined miRNA and mRNA signature identifies key molecular players and pathways involved in chikungunya virus infection in human cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fleming D.S., Miller L.C. Identification of small non-coding RNA classes expressed in swine whole blood during HP-PRRSV infection. Virology. 2018;517:56–61. doi: 10.1016/j.virol.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 66.Glazov E.A., Kongsuwan K., Assavalapsakul W., Horwood P.F., Mitter N., Mahony T.J. Repertoire of bovine miRNA and miRNA-like small regulatory RNAs expressed upon viral infection. PLoS One. 2009;4:e6349. doi: 10.1371/journal.pone.0006349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Juranic Lisnic V., Babic Cac M., Lisnic B., Trsan T., Mefferd A., Das Mukhopadhyay C., Cook C.H., Jonjic S., Trgovcich J. Dual analysis of the murine cytomegalovirus and host cell transcriptomes reveal new aspects of the virus-host cell interface. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hutzinger R., Feederle R., Mrazek J., Schiefermeier N., Balwierz P.J., Zavolan M., Polacek N., Delecluse H.J., Huttenhofer A. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eckwahl M.J., Sim S., Smith D., Telesnitsky A., Wolin S.L. A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway. Genes Dev. 2015;29:646–657. doi: 10.1101/gad.258731.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eckwahl M.J., Arnion H., Kharytonchyk S., Zang T., Bieniasz P.D., Telesnitsky A., Wolin S.L. Analysis of the human immunodeficiency virus-1 RNA packageome. RNA. 2016;22:1228–1238. doi: 10.1261/rna.057299.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rakitina D.V., Taliansky M., Brown J.W., Kalinina N.O. Two RNA-binding sites in plant fibrillarin provide interactions with various RNA substrates. Nucleic Acids Res. 2011;39:8869–8880. doi: 10.1093/nar/gkr594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoo D., Wootton S.K., Li G., Song C., Rowland R.R. Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA-associated protein fibrillarin. J. Virol. 2003;77:12173–12183. doi: 10.1128/JVI.77.22.12173-12183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Canetta E., Kim S.H., Kalinina N.O., Shaw J., Adya A.K., Gillespie T., Brown J.W., Taliansky M. A plant virus movement protein forms ringlike complexes with the major nucleolar protein, fibrillarin, in vitro. J. Mol. Biol. 2008;376:932–937. doi: 10.1016/j.jmb.2007.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deffrasnes C., Marsh G.A., Foo C.H., Rootes C.L., Gould C.M., Grusovin J., Monaghan P., Lo M.K., Tompkins S.M., Adams T.E. Genome-wide siRNA screening at biosafety level 4 reveals a crucial role for fibrillarin in henipavirus infection. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koonin E.V., Moss B. Viruses know more than one way to don a cap. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3283–3284. doi: 10.1073/pnas.0915061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Issur M., Geiss B.J., Bougie I., Picard-Jean F., Despins S., Mayette J., Hobdey S.E., Bisaillon M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009;15:2340–2350. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ogino T., Yadav S.P., Banerjee A.K. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3463–3468. doi: 10.1073/pnas.0913083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Plotch S.J., Bouloy M., Krug R.M. Transfer of 5'-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc. Natl. Acad. Sci. U. S. A. 1979;76:1618–1622. doi: 10.1073/pnas.76.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koppstein D., Ashour J., Bartel D.P. Sequencing the cap-snatching repertoire of H1N1 influenza provides insight into the mechanism of viral transcription initiation. Nucleic Acids Res. 2015;43:5052–5064. doi: 10.1093/nar/gkv333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gu W., Gallagher G.R., Dai W., Liu P., Li R., Trombly M.I., Gammon D.B., Mello C.C., Wang J.P., Finberg R.W. Influenza A virus preferentially snatches noncoding RNA caps. RNA. 2015;21:2067–2075. doi: 10.1261/rna.054221.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lukarska M., Fournier G., Pflug A., Resa-Infante P., Reich S., Naffakh N., Cusack S. Structural basis of an essential interaction between influenza polymerase and Pol II CTD. Nature. 2017;541:117–121. doi: 10.1038/nature20594. [DOI] [PubMed] [Google Scholar]
- 82.Beyer A.R., Bann D.V., Rice B., Pultz I.S., Kane M., Goff S.P., Golovkina T.V., Parent L.J. Nucleolar trafficking of the mouse mammary tumor virus gag protein induced by interaction with ribosomal protein L9. J. Virol. 2013;87:1069–1082. doi: 10.1128/JVI.02463-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Howard K., Cherezova L., DeMaster L.K., Rose T.M. ORF73 LANA homologs of RRV and MneRV2 contain an extended RGG/RG-rich nuclear and nucleolar localization signal that interacts directly with importin beta1 for non-classical nuclear import. Virology. 2017;511:152–164. doi: 10.1016/j.virol.2017.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Melen K., Tynell J., Fagerlund R., Roussel P., Hernandez-Verdun D., Julkunen I. Influenza A H3N2 subtype virus NS1 protein targets into the nucleus and binds primarily via its C-terminal NLS2/NoLS to nucleolin and fibrillarin. Virol. J. 2012;9:167. doi: 10.1186/1743-422X-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pentecost M., Vashisht A.A., Lester T., Voros T., Beaty S.M., Park A., Wang Y.E., Yun T.E., Freiberg A.N., Wohlschlegel J.A. Evidence for ubiquitin-regulated nuclear and subnuclear trafficking among Paramyxovirinae matrix proteins. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shi D., Lv M., Chen J., Shi H., Zhang S., Zhang X., Feng L. Molecular characterizations of subcellular localization signals in the nucleocapsid protein of porcine epidemic diarrhea virus. Viruses. 2014;6:1253–1273. doi: 10.3390/v6031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stewart M., Hardy A., Barry G., Pinto R.M., Caporale M., Melzi E., Hughes J., Taggart A., Janowicz A., Varela M. Characterization of a second open reading frame in genome segment 10 of bluetongue virus. J. Gen. Virol. 2015;96:3280–3293. doi: 10.1099/jgv.0.000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wulan W.N., Heydet D., Walker E.J., Gahan M.E., Ghildyal R. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front. Microbiol. 2015;6:553. doi: 10.3389/fmicb.2015.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marfori M., Mynott A., Ellis J.J., Mehdi A.M., Saunders N.F., Curmi P.M., Forwood J.K., Boden M., Kobe B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta. 2011;1813:1562–1577. doi: 10.1016/j.bbamcr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 90.Hiscox J.A. The nucleolus--a gateway to viral infection? Arch. Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hiscox J.A. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rawlinson S.M., Moseley G.W. The nucleolar interface of RNA viruses. Cell Microbiol. 2015;17:1108–1120. doi: 10.1111/cmi.12465. [DOI] [PubMed] [Google Scholar]
- 93.Salvetti A., Greco A. Viruses and the nucleolus: the fatal attraction. Biochim. Biophys. Acta. 2014;1842:840–847. doi: 10.1016/j.bbadis.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lindstrom M.S. NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem Res Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Passos-Castilho A.M., Marchand C., Archambault D. B23/nucleophosmin interacts with bovine immunodeficiency virus Rev protein and facilitates viral replication. Virology. 2018;515:158–164. doi: 10.1016/j.virol.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 96.Fankhauser C., Izaurralde E., Adachi Y., Wingfield P., Laemmli U.K. Specific complex of human immunodeficiency virus type 1 rev and nucleolar B23 proteins: dissociation by the Rev response element. Mol. Cell Biol. 1991;11:2567–2575. doi: 10.1128/mcb.11.5.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shi D., Shi H., Sun D., Chen J., Zhang X., Wang X., Zhang J., Ji Z., Liu J., Cao L. Nucleocapsid interacts with NPM1 and protects it from proteolytic cleavage, enhancing cell survival, and is involved in PEDV growth. Sci. Rep. 2017;7:39700. doi: 10.1038/srep39700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mai W., Huang F., Chen H., Zhou Y., Chen Y. Nervous necrosis virus capsid protein exploits nucleolar phosphoprotein Nucleophosmin (B23) function for viral replication. Virus Res. 2017;230:1–6. doi: 10.1016/j.virusres.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 99.Duan Z., Chen J., Xu H., Zhu J., Li Q., He L., Liu H., Hu S., Liu X. The nucleolar phosphoprotein B23 targets Newcastle disease virus matrix protein to the nucleoli and facilitates viral replication. Virology. 2014;452–453:212–222. doi: 10.1016/j.virol.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 100.Tajrishi M.M., Tuteja R., Tuteja N. Nucleolin: the most abundant multifunctional phosphoprotein of nucleolus. Commun. Integr. Biol. 2011;4:267–275. doi: 10.4161/cib.4.3.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Strang B.L., Boulant S., Kirchhausen T., Coen D.M. Host cell nucleolin is required to maintain the architecture of human cytomegalovirus replication compartments. mBio. 2012;3 doi: 10.1128/mBio.00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]