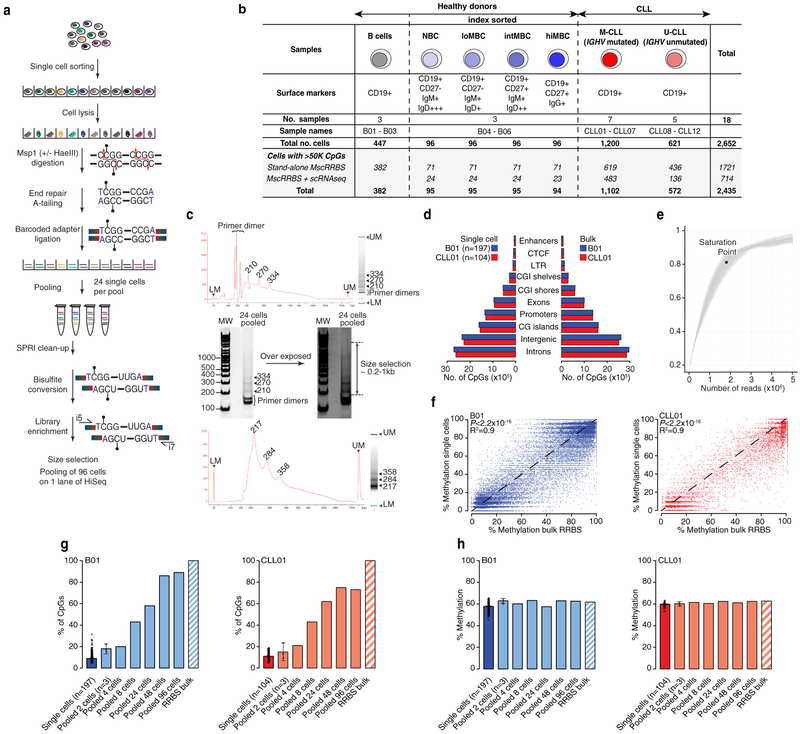
Extended Data Figure 1. MscRRBS is an accurate and reproducible method for single-cell DNAme analysis.
(a) Detailed schematic of multiplexed single-cell RRBS (MscRRBS) protocol. (b) Extended summary table of healthy donors and CLL patient samples used in this study. (c) Representative size distribution of the MscRRBS libraries assessed by Agilent Bioanalyzer before and after primer dimers removal. The DNA fragment size in MscRRBS libraries is typically 200–1000 bp, with some visible peaks corresponding to the MspI fragments for repeat elements, and primer dimer contaminants (~170 bp). (d) Number of CpGs observed in MscRRBS libraries across relevant genomic regions comparing MscRRBS (left) and bulk RRBS (right) assays for normal B (B01) and CLL (CLL01) cells. The enrichment in exons, promoters and CpG islands (CGIs) observed in MscRRBS libraries corresponded to ~40% of the total sequenced CpGs, akin to bulk RRBS assays. (e) Downsampling analysis showing that ~1.7 million paired-end reads per cell provided ~85% of unique CpGs with further sequencing resulting in a marginal increase in coverage. (f) Correlation of average CpG methylation across in silico merged single cells and bulk RRBS obtained from matched samples for normal B (B01, n = 40,257 CpGs) and CLL (CLL01, n = 9,578 CpGs) cells. P-values are indicated for two-sided Pearson’s correlation test. (g) Pooling individual single cells together rapidly increases the number of CpGs recovered, approaching bulk RRBS coverage with >48 cells. The percentage of CpG sites detected in single cell data (blue and red for normal B cells and CLL cells, respectively), the in vitro pooled single-cell datasets (light blue and light red, respectively) and matched bulk RRBS libraries (striped bars) is shown. Error bars represent 95% confidence interval. (h) Same as panel (g) for average CpG DNA methylation. Single, pooled cells and bulk RRBS showed a similar CpG methylation percentage, suggesting measured genome-wide DNAme profiles of individual cells accurately recapitulate bulk methylation profiles in the same cell type.