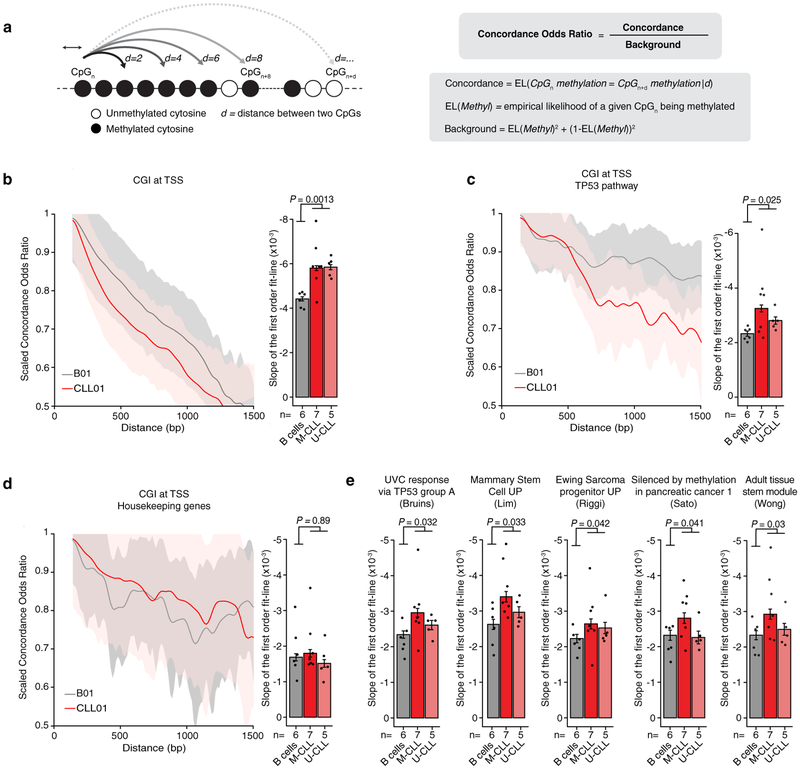
Extended Data Figure 4. Long-range DNA methylation concordance decay.
(a) Concordance odds ratio (COR) of DNA methylation state between any two neighbouring CpGs as function of their genomic distance (see Methods for details). (b) Left: Scaled COR (0–1) for CpG islands at transcription start sites (CGI at TSS; B01 and CLL01 samples are shown as a representative example). Right: average rate of decay (slope of the first order fit line) in COR for normal B (n = 6) and CLL (n = 12) samples for CGI at TSS (B01-B06 [n = 666 cells; n = 48,065,000 CpGs] and CLL01-CLL12 [M-CLL, n = 619 cells, n = 38,968,846 CpGs; U-CLL, n = 436 cells, n = 37,464,310 CpGs]). P-value was computed for two-sided Mann-Whitney U-test by comparing the average rate of decay in COR of healthy donor samples (n = 6) with the average rate of decay in COR of CLL samples (n = 12). (c) Same as panel (b) for CGI at TSS of genes belonging to the TP53 gene set69. Normal B cells, n = 666 cells, n = 6,308,174 CpGs; M-CLL, n = 619 cells, n = 5,113,493 CpGs; U-CLL, n = 436 cells, n = 4,982,039 CpGs. P-value was computed for two-sided Mann-Whitney U-test by comparing the average rate of decay in COR of healthy donor samples (n = 6) with the average rate of decay in COR of CLL samples (n = 12). (d) Same as panel (b) for CGI at TSS of housekeeping genes70. Normal B cells, n = 666 cells, n = 2,087,432 CpGs; M-CLL, n = 619 cells, n = 1,686,295 CpGs; U-CLL, n = 436 cells, n = 1,620,802 CpGs. P-value was computed for two-sided Mann-Whitney U-test by comparing the average rate of decay in COR of healthy donor samples (n = 6) with the average rate of decay in COR of CLL samples (n = 12). (e) Average rate of decay in COR for normal B (n = 6) and CLL (n = 12) samples for CGI at TSS of genes belonging to gene sets previously reported to be affected by high epimutation rate6. Normal B cells, n = 666 cells, n = 48,065,000 CpGs; M-CLL, n = 619 cells, n = 38,968,846 CpGs; U-CLL, n = 436 cells, n = 37,464,310 CpGs. P-value was computed for two-sided Mann-Whitney U-test by comparing the average rate of decay in COR of healthy donor samples (n = 6) with the average rate of decay in COR of CLL samples (n = 12). Throughout the figure, error bars represent 95% confidence interval.