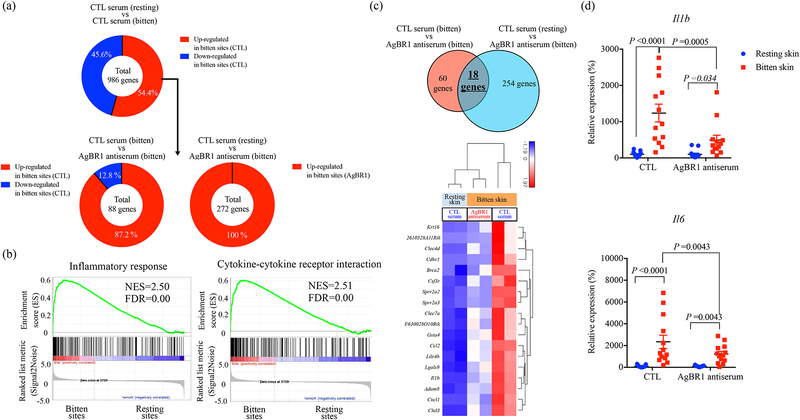
Figure 4. AgBR1 antiserum modulates host responses at the mosquito bite site.
(a) (Top panel) 536 genes (54.4 %) within 986 differentially expressed genes (P < 0.05) were upregulated at the bitten sites of mice administered control serum. (Bottom left panel) Among these 536 genes, 78 genes were significantly upregulated at the bitten site of mice administered control serum compared with mice injected with AgBR1 antiserum. (Bottom right panel) Among the 536 genes, 272 genes were differentially upregulated in bitten sites of mice administered AgBR1 antiserum compared with the resting sites of mice inoculated with control serum. (b) GSEA of inflammatory responses (Hallmark) and cytokine-cytokine receptor interaction (KEGG) pathway enriched at bite sites of mice compared with resting sites in control mice. NES, normalized enrichment score. (c) (Top panel) Venn diagram depicting the overlap of genes differentially expressed across the conditions. (Bottom panel) Heat map of hierarchical clustering performed on 18 upregulated genes across the conditions (Fold change >1.5, P < 0.05). (a-c) Control-resting skin: n=2, Control-bitten skin: n=2, AgBR1 antiserum-bitten skin: n=2 biologically independent samples. Normalized read counts were statistically modeled using Partek Flow’s Gene Specific Analysis (GSA) approach. (d) QRT-PCR based analysis of Il1b and Il6 expression, which is normalized to mouse β actin RNA levels. Each dot represents one bitten or control site. Data are presented as mean ± s.e.m.. Significance was determined by two-way ANOVA test. (Control-resting skin: n=13, Control-bitten skin: n=13, AgBR1 antiserum-resting skin: n=13, AgBR1 antiserum-bitten skin: n=13 biologically independent samples pooled from two separate experiments.)