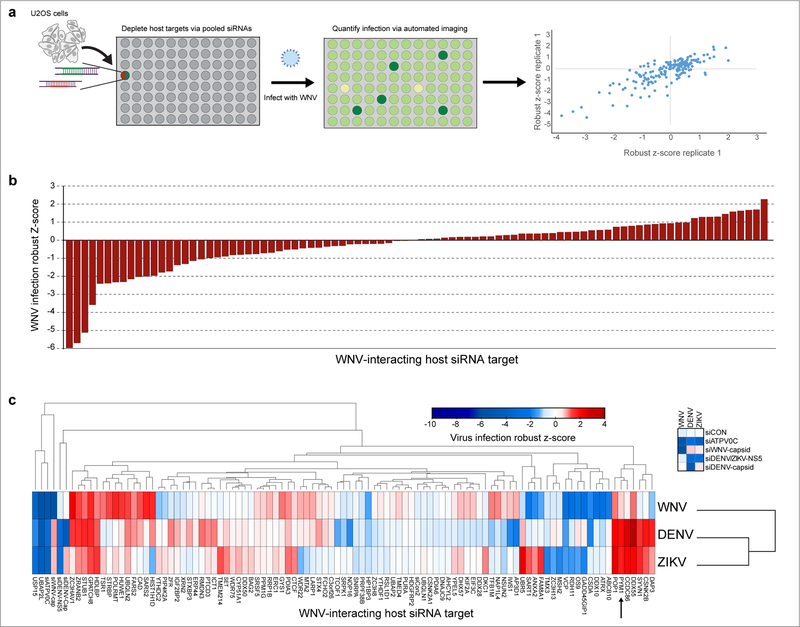
Fig. 4. RNAi screening reveals important roles for host proteins that interact with WNV proteins.
a, Schematic of the WNV infection siRNA screen. 122 WNV-interacting host proteins were depleted using a pool of 2 siRNAs for each target (x-axis). The screen was performed with two independent biological replicates and robust z-scores of WNV infection are plotted for each individual replicate. b, The average robust Z-score for each of the 88 (non-toxic) genes calculated from two independent biological replicates is shown. c, siRNA screening against WNV, DENV, and ZIKV performed with two independent biological experiments. The colors in the heatmap correspond to the average robust z-score of infection and are indicated in the accompanying scale. Columns (viruses) were clustered using One minus Pearson correlation and rows (targets) were clustered by Euclidean distance. The WNV capsid-interacting host protein PYM1 is indicated with an arrow. The complete screening dataset is provided in Supplementary Table 8.