Figure 1.
Design of a Chimpanzee Env-Stabilized Trimer and Binding to V2-Apex bnAb iGL Abs
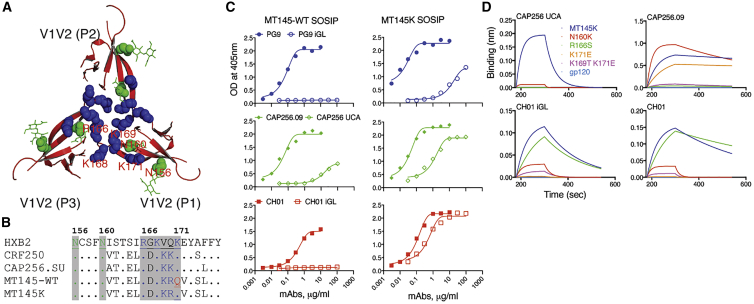
(A) Structural arrangement of the V2-apex bnAb core epitope region on the BG505.664 soluble Env trimer (modified from Garces et al., 2015; PDB: 5CEZ). The ribbon representation of V1V2 loop strands that form the trimer apex show a cluster of positively charged lysine-rich peptide regions (HXB2-R166-K171: R or K residues shown as blue spheres) and the two glycans N156 and N160 (depicted in green spheres with lines). The side chains of the positively charged residues intersperse with the side chains of residues from adjacent protomers to form a continuous positively charged surface at the tip of the trimer to provide a minimal V2-apex bnAb epitope.
(B) Amino-acid sequence alignment of strand B and C V2 of HIV CRF250, CAP256.SU, chimpanzee SIV MT145 WT, and its V2-modified variant (Q171K), MT145K. Glutamine (Q) at position 171 (shown in red) was substituted with lysine (K) in MT145 Env to gain binding to V2-apex bnAb inferred germline (iGL) Abs.
(C) ELISA binding of mature V2-apex bnAbs, PG9, CAP256.09, and CH01 and their iGL versions to WT MT145 (red) and MT145K SOSIP trimers.
(D) Octet binding curves (association, 120 s [180–300]; dissociation, 240 s [300–540]) of CAP256 UCA and CH01 iGL Abs and their respective mature Ab versions (CAP256.09 and CH01) to MT145K trimer, its glycan knockout (N160K) variant, K-rich core epitope substituted variants, and the corresponding monomeric gp120. The Abs were immobilized on human IgG Fc capture biosensors and 1uM trimer or gp120 proteins used as analytes. The binding response is shown in nanometers (nm).