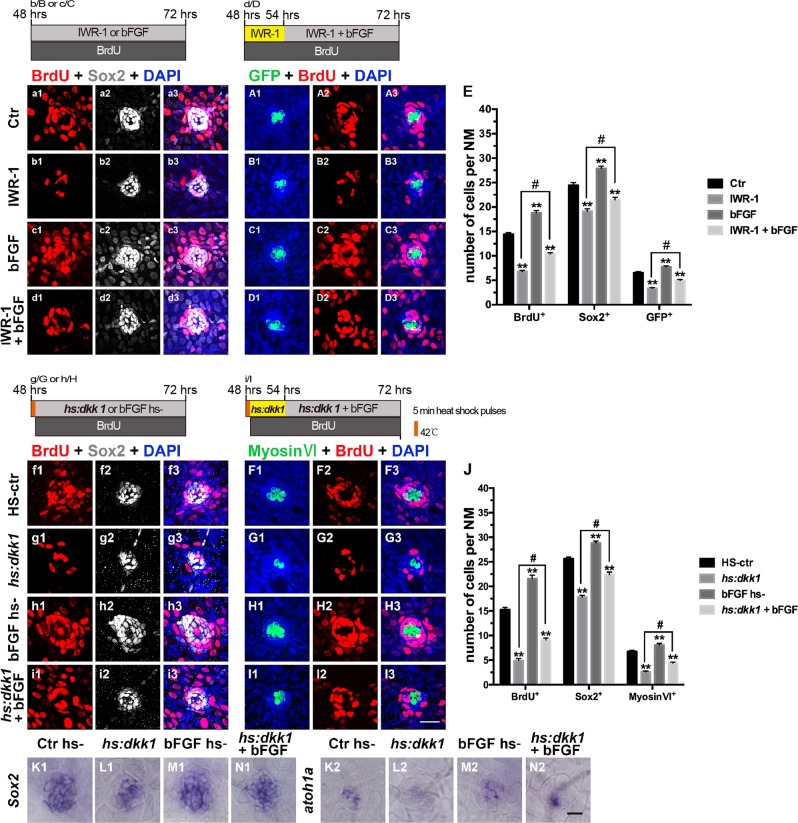
Fig. 4. Effects of FGF activation on cell proliferation in Wnt-inhibited larvae.
a1–D3: Proliferative cells labeled with BrdU (red), SCs labeled with Sox2 (white), and HCs labeled with GFP (green). a1–A3: Control neuromast; b1–B3: After addition of IWR–1; c1–C3: After addition of bFGF; d1–D3: After 6 h of incubation with IWR–1, bFGF and IWR–1 were added for 18 h; e: Quantification of the proliferative cells (BrdU+), SCs (Sox2+), and HCs (GFP+) shown in a1–D3. f1–I3: Proliferative cells labeled with BrdU (red), SCs labeled with Sox2 (white), and HCs labeled with myosin VI (green). f1–F3: Heat shock-negative control larvae (HS–ctr); g1–G3: Wnt repression by heat shock induction of the hs:dkk1 transgene (hs:dkk1); h1–H3: After addition of bFGF; i1–I3: At 54 hpf, hs:dkk1 embryos were treated with bFGF for 18 h. j: Quantification of the proliferative cells (BrdU+), SCs (Sox2+), and HCs (myosin VI+) shown in f1–I3. n = 5–7 fish per group. ** Indicates p < 0.001, # indicates p < 0.05, and error bars indicate the standard error of the mean. K1–N1 and K2–N2: WISH analysis of Sox2 (K1–N1) and atoh1a (K2–N2) expression in the neuromasts from different groups. Scale bar in I3 = 20 μm for a1–D3 and f1–I3. Scale bar in N2 = 30 μm for K1–N1, K2–N2