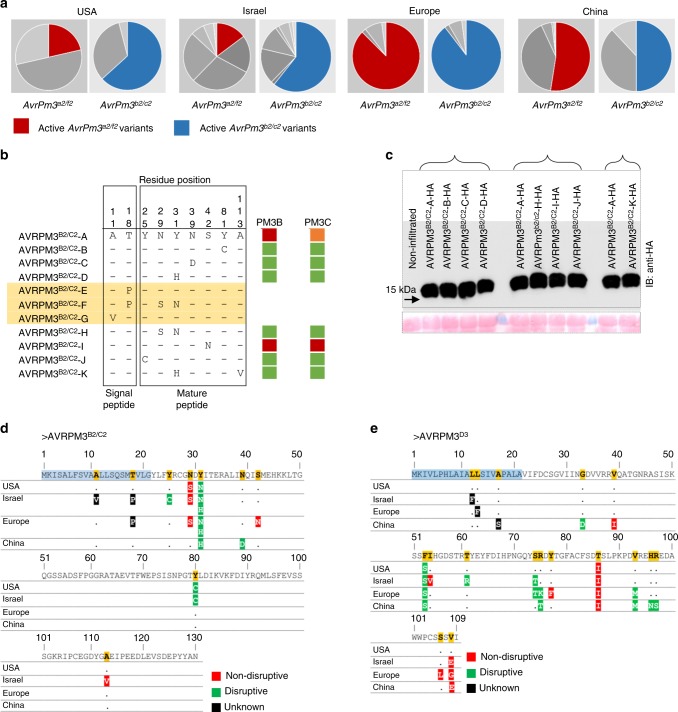
Fig. 4.
Functional characterisation of AVRPM3B2/C2 and AVRPM3D3 based on natural and artificial sequence variation. a Geographical distribution and frequency of AvrPm3b2/c2 sequence haplotypes encoding active AVRPM3B2/C2 virulence protein variant as compared to AvrPm3a2/f2. Underlying source data is provided in a Source Data File. b Summary of AVRPM3B2/C2 haplovariants, only polymorphic residues are depicted. Strong recognition by PM3B and PM3C based on N. benthamiana infiltration assays are indicated with a red box, and weak recognition with an orange box. All assays are based on expression of the mature protein after removal of the signal peptide. Therefore, haplovariants E and G, and haplovariant F were not tested because the mature protein sequence is identical to variant A and variant H, respectively. c Western blot detection (upper panel) of C terminal HA epitope fusion to the mature proteins of the AVRPM3B2/C2 haplovariants. Ponceau staining of the western blot membranes is depicted in the lower panel. Braces indicate samples where all constructs were combined on the same leaf and rotated together with AVRPM3B2/C2-A as a reference. Uncropped Western blot images are provided in a Source Data File. d, e Functional characterisation of the impact of natural sequence diversity of AVRPM3B2/C2 (d) and AVRPM3D3 (e) on AVR function