Figure 2.
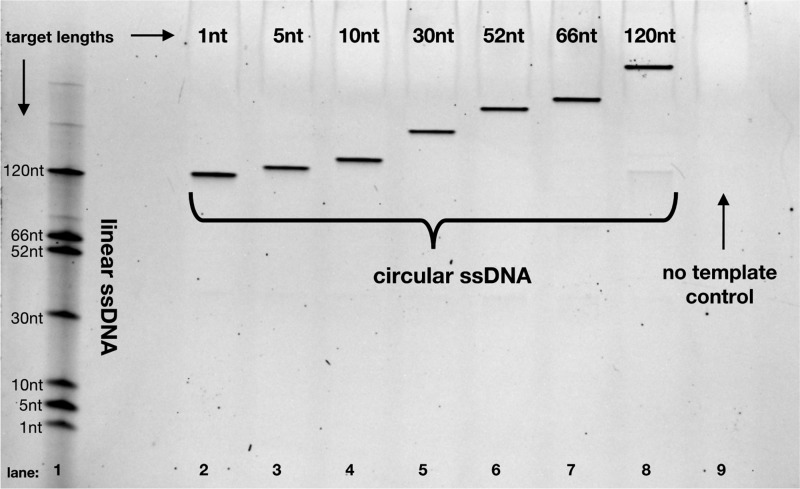
Circularization can be performed on target sequences as short as a single nucleotide with reaction efficiency that is independent of target sequence length. After five rounds of temperature cycling and subsequent exonuclease treatment, we achieve consistently efficient circularization for target sequences ranging in length from 1 to 120 nt (lanes 2–8). In this denaturing gel, lane 1 contains a mixture of all the linear ssDNA target sequences. The lengths listed are the lengths of the target region; the full lengths in lane 1 have additional flanking 28- and 23-nt anchor sites, and the full lengths in lanes 2–8 have an additional 102 nt from the MIP. Lane 9 illustrates that no circular DNA is produced in the absence of the target sequence.