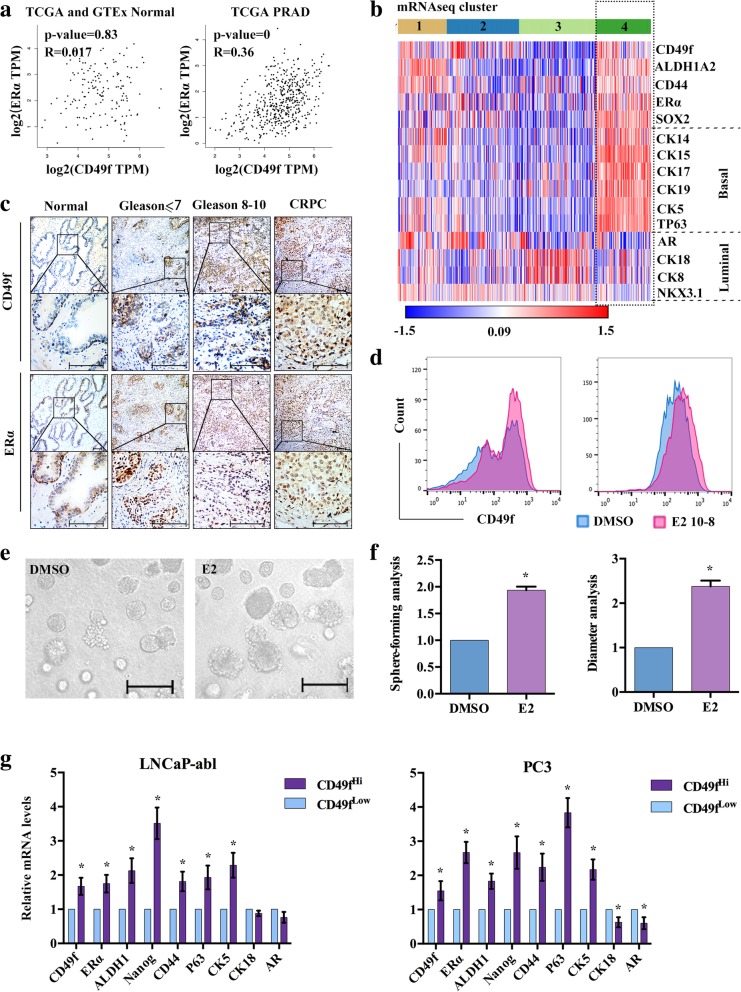
Fig. 1.
The expression of CD49f and ERα. a, The correlation of CD49f and ERα in normal prostate and PRAD patients from TCGA (num (Normal) = 52), num (PRAD) = 498)) and GTEx (num (Normal) = 100) datasets. b, Heat map analysis of the differentially expressed genes in PRAD patients. The TCGA consortium devised a subclassification of prostate cancers (num(N) = 497) into four distinct groups (1–4) based upon mRNA-Seq clustering and z-score analysis in Morpheus with a P ≤ 0.05 criterion, Rows: samples; columns: the indicated genes. c, IHC staining of CD49f and ERα in normal prostate tissues and cancer tissues with different Gleason score. Scale bar, 200 μm. d, Flow cytometry analysis of the expression of CD49f in LNCaP-abl and PC3 treated with either DMSO or 10 nM E2 for 72 h (n = 3). e, Analysis of enriching stem cell spheres treated with E2 for 2 weeks. Scale bar, 100 μm. f, Sphere-forming and diameter analysis of e. g, Flow cytometry sorting of CD49fHi and CD49fLow cells in LNCaP-abl and PC3, qRT-PCR analysis showing expression changes of the indicated genes. The data are presented as the means±SD (n = 3). *, P < 0.05 vs. CD49fLow PCBSLCs. Abbreviate: PRAD: Prostate Adenocarcinoma