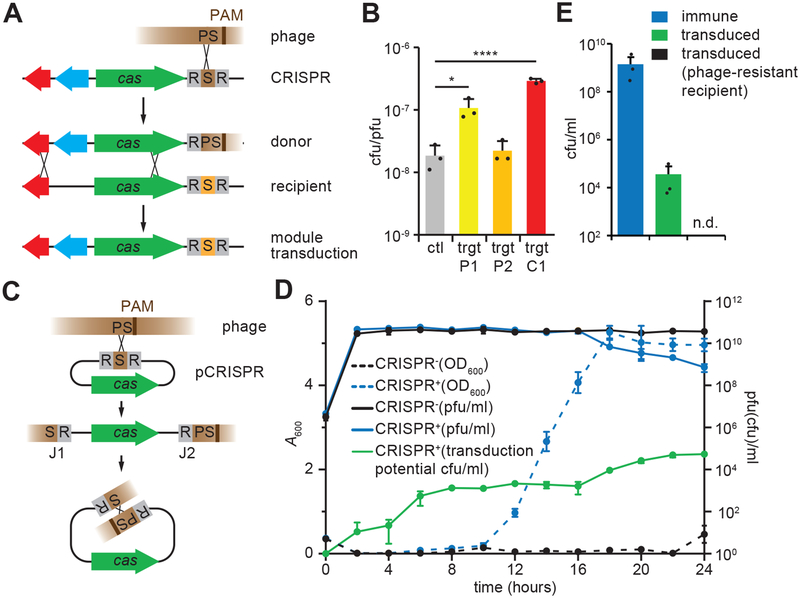
Figure 1. Transfer of CRISPR-Cas elements through spacer-mediated transduction.
(A) Transduction of CRISPR-adjacent loci (blue arrow) after phage recombination with a chromosomal CRISPR-cas locus; “S” CRISPR spacer, “PS” protospacer. (B) Transducing particle production from S. aureus strain 08BA02176 tagged with an erythromycin resistance cassette. Liquid cultures were infected at a MOI (multiplicity of infection) of 50. Wild-type phage (ctl) or phage with the target site of the first spacer of the CRISPR array inserted at 15 kb (P1) or 20 kb (P2) positions on the phage genome were used. Wild-type phage was also used to infect a strain with a chromosomally inserted phage-targeting spacer (C1). Mean + STD of 3 biological replicates are reported. Two-tailed unpaired t-test was used to calculate P value, *p = 0.023, ****p = 0.00058 (C) Transduction of a plasmid containing a CRISPR-cas locus after recombination with the phage genome. J1 and J2, recombination junctions. (D) Growth and phage titers of infected cultures of bacteria containing plasmids with either the type II-A CRISPR-Cas system (CRISPR+) or the empty vector control (CRISPR−). Liquid cultures were infected at a MOI of 1 with ΦNM4γ4. The growth of cultures was determined by measurement of absorbance at 600 nm (A600). Titers, plaque forming units/ml (pfu/ml), and levels of transducing-immune phage particles, colony forming units/ml (cfu/ml), were determined every time point. No transducing-immune particles were detected using a vector control. Mean ± STD of 3 biological replicates are reported. (E) Levels of transduction during CRISPR adaption were determined by mixing cells at a 1 CRISPR+ (naïve, without a targeting spacer) to 5 CRISPR− ratio and infecting the mix with ΦNM4γ4 at an MOI of 1. Cultures were collected 20 hours post infection and survivors resulting from the acquisition of new spacers or the transduction of the adapted pCRISPR were measured by enumerating colonies on plates containing different antibiotic combinations. As a control, the experiment was repeated after mixing non-CRISPR resistant cells with naïve CRISPR+ staphylococci. Mean ± STD of 3 biological replicates are reported.