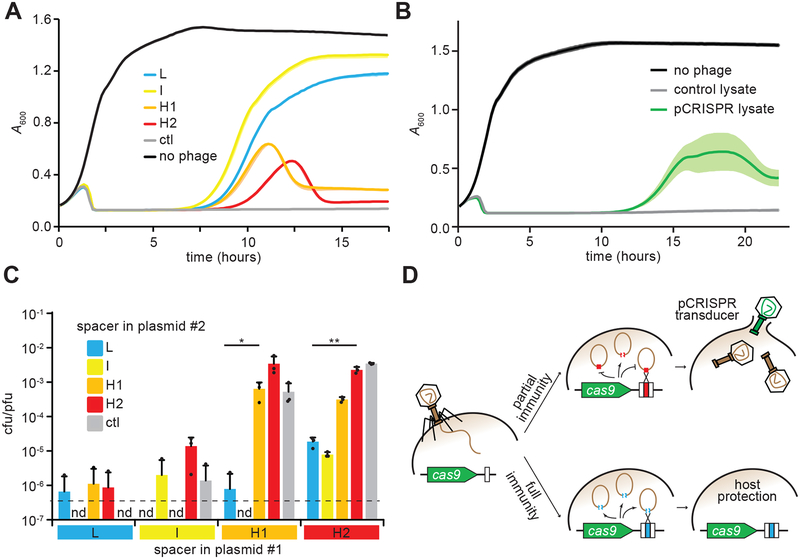
Figure 3. Spacers that mediate high pCRISPR transduction provide poor immunity to the host.
(A) Cell survival of cultures containing pCas9 with spacers L, I, H1 or H2, monitored as growth after infection with ΦNM4γ4 at a MOI of 1. No spacer (ctl) and no phage experiments were used as controls. Growth was recorded by following the absorbance at 600 nm (A600) of the cultures. Mean - STD of 3 biological replicates are reported. (B) Cell survival of cultures lacking CRISPR-Cas immunity, monitored as growth after infection with a cell lysate obtained after infection of staphylococci harboring pCRISPR (or an empty vector as a control) with ΦNM4γ4 at a MOI of 1. This lysate contains both phage as well as plasmid transducing plasmids. Growth was recorded by following the absorbance at 600 nm (A600) of the cultures. Mean ± STD of 5 biological replicates are reported. (C) Transduction efficiency of plasmid #2, measured as transductant cfu / total pfu present in lysates of staphylococci harboring two pCas9 plasmids with different spacers, collected 90 minutes after infection with ΦNM4γ4 at a MOI of 50. Limit of detection is 1.5 cfu/ml (dotted line). Mean + STD of 3 biological replicates are reported. Two-tailed unpaired t-test was used to calculate P values, *p = 0.034, **p = 0.0012 (D) Model for spacer-mediated p CRISPR transduction. During the type II-A CRISPR-Cas immune response, efficient spacers fully protect the host and prevent the generation of both phage progeny and transducing particles. In contrast, spacers that mediate poor immunity allow the lysis of some of the infected hosts, enabling the release of pCRISPR transducing particles generated after the recombination of the plasmid and the phage genome at the spacer/protospacer sequences.