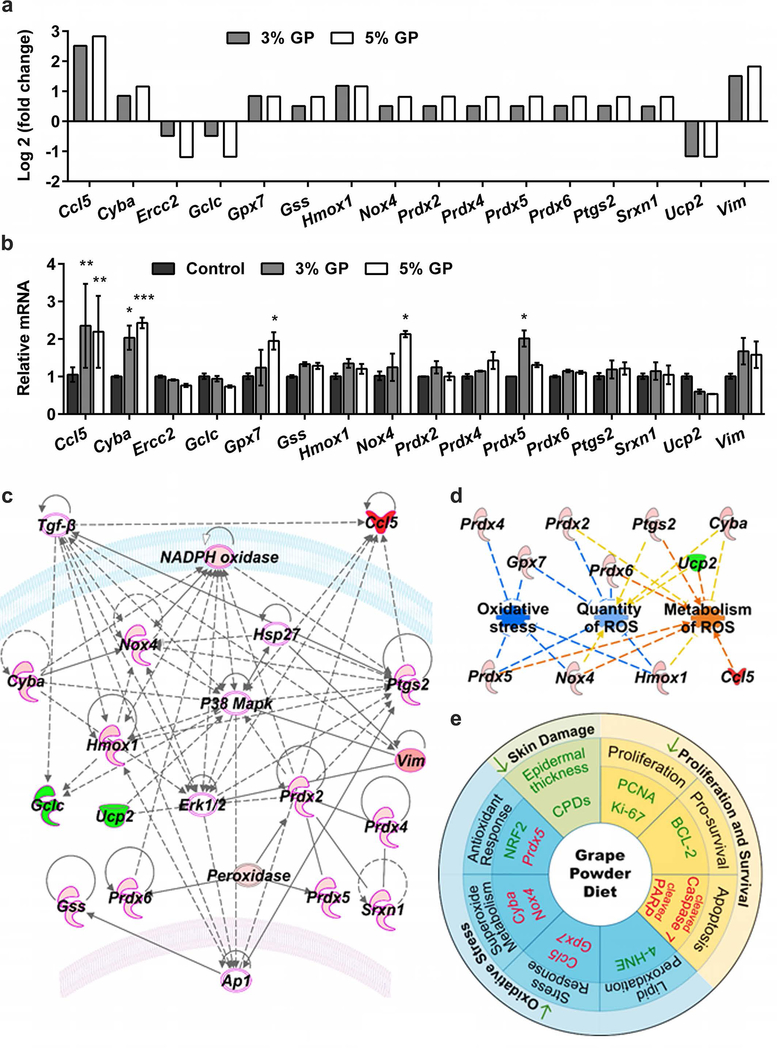
Figure 6. Mechanistic analysis of GP-mediated chemoprotective effects against UVB-induced skin carcinogenesis.
(a) Differentially expressed genes in 3 and 5% GP group from the oxidative stress PCR array. Genes with ≥1.75-fold change and p<0.05 in response to 5% GP and ≥1.4-fold change in response to 3% GP are included. P-values are calculated based on Student’s t-test of the replicate 2^(-ΔCT) values for each gene. (b) RT-qPCR validation of key altered genes in response to 3 and 5% GP. The data presented here are from RNA pooled evenly from 9 mouse tumor samples per group as described in materials and methods. Statistical analysis performed using two-way ANOVA with Tukey’s multiple comparison. Data are represented as mean ± SEM (*p<0.05, **p<0.01, ***p<0.001). (c) A gene network pathway was generated with key altered genes (≥1.75-fold change in response to 5% GP) using IPA software. Red= upregulated genes; Green= downregulated genes; Pink outline= Cancer-associated genes; Uncolored nodes= genes not included in the PCR array but found during IPA analysis. Gene-gene interactions are indicated by arrows, the solid lines denote a robust correlation with partner genes, and dashed lines indicate statistically significant but less frequent correlations. (d) Using IPA, the functional annotations of 5% GP-modulated genes were generated showing inhibition of oxidative stress and quantity of ROS, as well as increased metabolism of ROS. Blue lines= inhibition; Orange= activation; Yellow= findings inconsistent with the state of downstream molecules. (e) Schematic representation of the effects of dietary GP on skin tumorigenesis are presented. Green= downregulated/decreased, Red= upregulated/increased measurements.