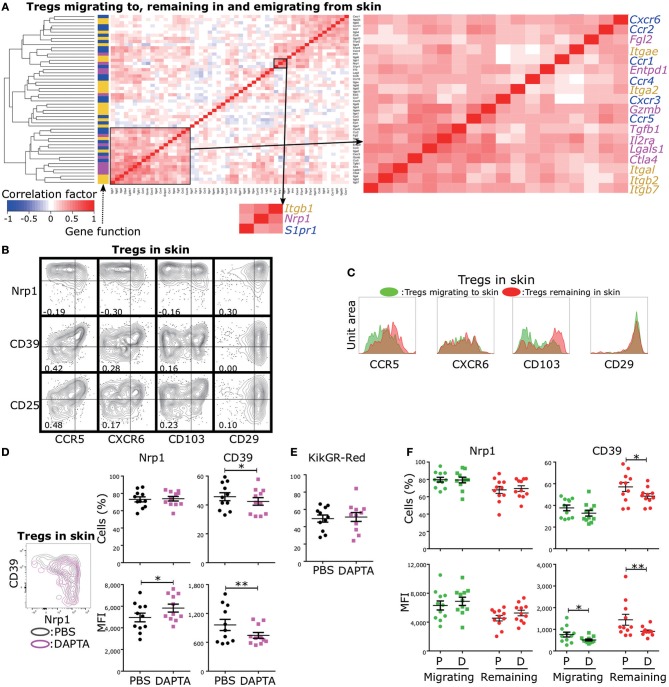
Figure 5.
Identification of migration-related molecules influencing functional phenotypic diversity of skin Tregs. (A) Cross-correlation analysis of the expression profiles of genes related to Treg function (magenta), migration (blue), and adhesion (yellow). scqPCR data were formed by concatenating the data of the three Treg populations (Tregs migrating to, remaining in, and emigrating from skin). Colors in cells indicate positive (red) or negative (blue) correlations. Clustering was performed by the Manhattan distance and group average method. (B) Nrp1, CD39, CD25, CD29, CD103, CXCR6, and CCR5 protein expression by Tregs in skin. FCM data are representative of three independent experiments. Lines in plots indicate boundaries between positive and negative cells. (C) Replicative data of CD29, CD103, CXCR6, and CCR5 protein expression by Tregs migrating to and remaining in skin (n = 3). (D,E) Nrp1 and CD39 expression (D) and Proportions of KikGR-Red+ cells (E) in skin Tregs of mice treated with PBS (control) or DAPTA. (F) Nrp1 and CD39 expression by Tregs migrating to and remaining in skin of mice treated with PBS or DAPTA.