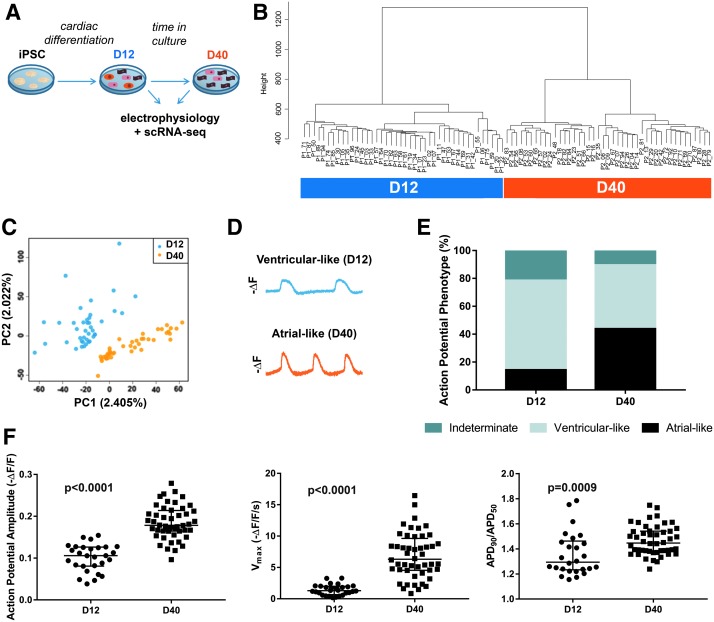
FIG. 3.
Single-cell RNA-seq analysis in hiPSC-derived cardiac differentiation clearly separates days 12 and 40 cells with distinct electrophysiological properties. (A) Schematic of experimental design. hiPSC-CMs were analyzed at days 12 and 40 of differentiation via single-cell RNA-seq and ArcLight. (B) Unsupervised hierarchical clustering on the entire transcriptomic profiles of all cells (D12: n = 42 cells; D40: n = 43 cells). (C) Principal component analysis compiled from the entire transcriptomic profiles of all cells. (D) Representative inverted fluorescent traces for ventricular-like morphology (from D12) and atrial-like morphology (from D40). (E) Percentage of total cells at D12 or D40 with each AP morphology classification. “Indeterminate” cells had either insufficiently mature APs or demonstrated a mixture of atrial- and ventricular-like APs. (F) AP amplitude (left), Vmax (middle), and APD90/APD50 (right) evaluated from D12 and D40 populations before submission for sequencing. P values were calculated by either a Student's t-test or Mann–Whitney U test. Data are reported as median ± interquartile range. hiPSC, human induced pluripotent stem cell.