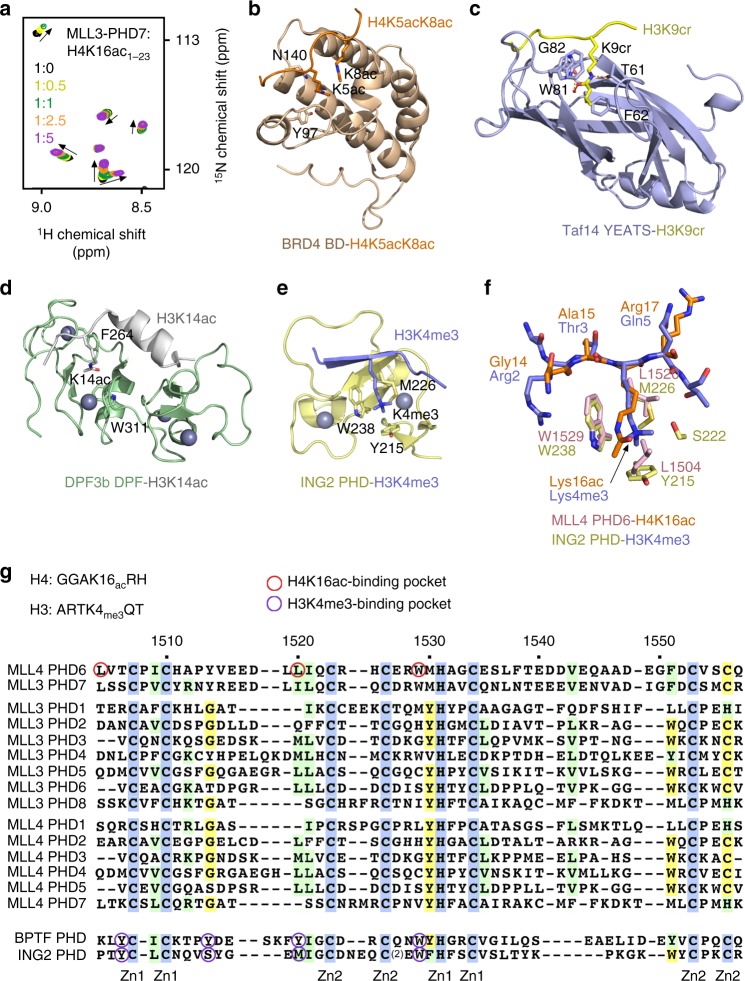
Fig. 7.
Comparative analysis of the H3K4me3 and H4K16ac recognition by histone readers. a Superimposed 1H,15N heteronuclear single quantum coherence spectra of MLL3-PHD71083–1143 collected upon titration with H4K16ac peptide. Spectra are color coded according to the protein:peptide molar ratio. b–d Structures of the BRD4 BD, Taf14 YEATS, and DPF3b DPF readers in complex with the indicated histone peptides24,50,51. PDB IDs: 3UVW (b), 5IOK (c), and 5I3l (d). e Structure of the ING2 PHD finger in complex with H3K4me330. PDB ID: 2G6Q. f A zoom-in view of the overlaid histone tail-binding sites of the ING2 PHD-H3K4me3 and MLL4 PHD6-H4K16ac complexes. g Alignment of amino acid sequences of the PHD fingers from human MLL4, MLL3, BPTF, and ING2. Absolutely, moderately, and weakly conserved residues are colored blue, green, and yellow, respectively. Zinc-coordinating cysteine/histidine residues are labeled