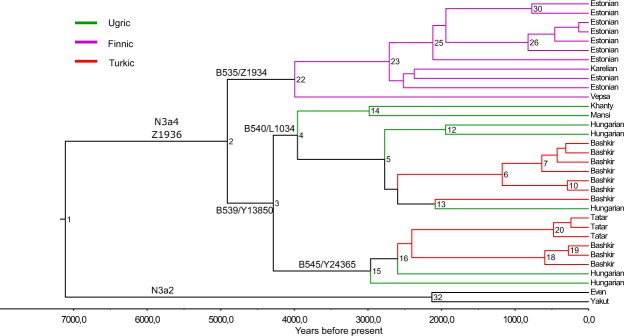
Figure 2.
Phylogenetic tree of hg N3a4. Phylogenetic tree of 33 high coverage Y-chromosomes from haplogroup N3a4 was reconstructed with BEAST v.1.7.5 software package. We used 8 sequences published in Karmin et al.63, 6 sequences published in Ilumäe et al.27, 2 sequences published in Wong et al.64 and 17 new sequences from this study. Two N3a2 samples were used as an outgroup to estimate coalescent times. Internal node numbers on the branches (not including nodes with low posterior values), sub-clade names and population names on the tips are indicated. Branches are coloured according to language affiliations. Number of branch-defining mutations and marker names are presented in Supplementary Fig. S4. All SNPs characterizing the clades (nodes) are presented in Supplementary Table S1. Age estimates can be found in Supplementary Table S2.