Fig. 1.
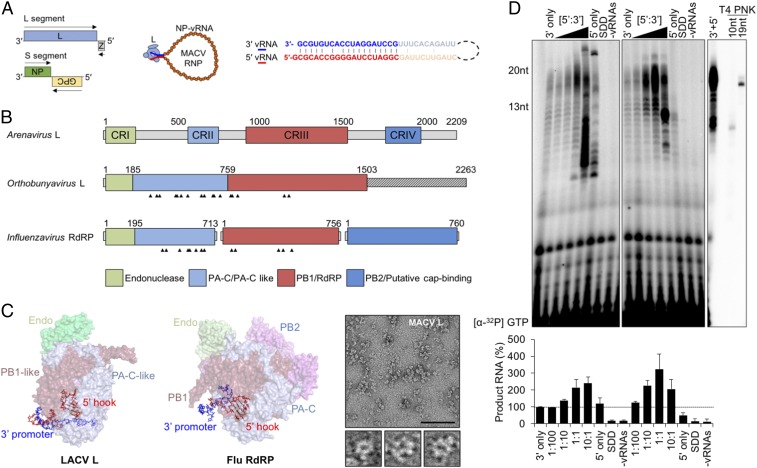
The 5′ genomic termini regulate the functions of SNS RNA polymerases. (A) The bisegmented ambisense arenavirus genome and the viral ribonucleoprotein (RNP), showing NP-encapsidated vRNA (orange) bound by L (light blue) with pseudocomplementary termini (bolded blue and red text, respectively). Viral ORFs are shown as colored boxes in the genome, and the arrows indicate coding sense. (B) Domain organization of the SNS RdRP. Four conserved regions (CRI to IV) of the arenavirus L protein are illustrated in colored boxes. The Orthobunyavirus and Influenzavirus domain labels are based on available crystal structures. The 5′ vRNA-interacting residues are indicated with arrowheads. (C) Positions of the 5′ vRNA hook-like structures in LACV L (Left, PDB ID code 5AMQ) and influenza A polymerase (Middle, Flu RdRP, PDB ID code 4WSB). The domains are colored according to the illustration in A. (C, Right) Visualization of purified MACV L by negative-stain electron microscopy. Three particles resembling the previously described L architecture (20) are shown below the larger micrograph image. (Scale bar: 100 nm.) (D) Stimulation of in vitro RNA synthesis by the 5′ vRNA. (D, Left) Increasing amounts of the 20-nt 5′ vRNA ligand were added to reactions relative to the 3′ template ([5′:3′]). Reactions with catalytically inactive L (SDD) or no added vRNA (−vRNAs) are shown. (D, Middle) Parallel reactions performed using a minimal activating ligand for RNA synthesis. (D, Right) A reaction performed in the presence of the 13-nt 5′ vRNA, alongside oligoribonucleotide primers end labeled by T4 polynucleotide kinase (PNK). Products of RNA synthesis were separated on denaturing 20% polyacrylamide-urea gels, visualized, and quantified as described in Materials and Methods, and % RNA synthesis is depicted in bar graphs. Error bars represent the SD from the mean of three independent experiments.