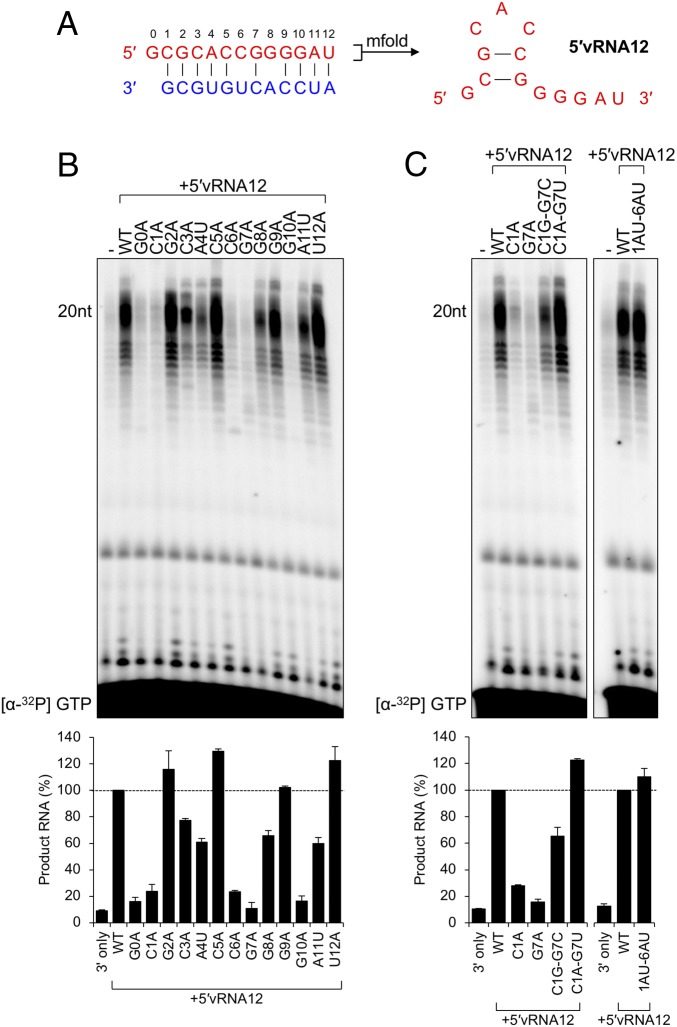
Fig. 4.
Identification of essential residues for a functional activating 5′vRNA12 ligand. (A) Predicted structure of the terminal 5′ vRNA ligand of MACV, determined by mfold as described in Materials and Methods. (B) Adenine/uracil mutagenesis of the MACV 12-nt 5′ vRNA ligand (5′vRNA12). In vitro RNA synthesis reactions were performed as in Fig. 2 with equimolar concentrations of 3′ promoter and 5′ ligand. Products of RNA synthesis were visualized and quantified as in Fig. 1, and the relative changes in RNA synthesis are shown in a bar graph below the gel image. (C) Substitution of predicted base-pairing events restores 5′ ligand functionality. The Left shows L-synthesized products for substitutions at the C1–G7 predicted base-pairing site. The Right shows the substitution of positions 1 to 2 and 6 to 7 with A-U to restore the predicted hook-like structure. All sequence names correspond to the mutant vRNAs described in SI Appendix, Table S2. Products of RNA synthesis were visualized and quantified as in Fig. 1, and the relative changes in RNA accumulation are shown in bar graphs below each gel image.