Significance
Mycobacterium tuberculosis (Mtb) causes the disease tuberculosis (TB), which kills more people than any other infection. The emergence of drug-resistant Mtb strains has exacerbated this already alarming epidemic. We have identified a small molecule, C10, that potentiates the activity of the frontline antibiotic isoniazid (INH) and prevents the selection for INH-resistant mutants. We find that C10 can even reverse INH resistance in Mtb. Therefore, our study reveals vulnerabilities that can be exploited to reverse INH resistance in Mtb.
Keywords: Mycobacterium tuberculosis, drug tolerance, antibiotic resistance, isoniazid, respiration
Abstract
Mycobacterium tuberculosis (Mtb) killed more people in 2017 than any other single infectious agent. This dangerous pathogen is able to withstand stresses imposed by the immune system and tolerate exposure to antibiotics, resulting in persistent infection. The global tuberculosis (TB) epidemic has been exacerbated by the emergence of mutant strains of Mtb that are resistant to frontline antibiotics. Thus, both phenotypic drug tolerance and genetic drug resistance are major obstacles to successful TB therapy. Using a chemical approach to identify compounds that block stress and drug tolerance, as opposed to traditional screens for compounds that kill Mtb, we identified a small molecule, C10, that blocks tolerance to oxidative stress, acid stress, and the frontline antibiotic isoniazid (INH). In addition, we found that C10 prevents the selection for INH-resistant mutants and restores INH sensitivity in otherwise INH-resistant Mtb strains harboring mutations in the katG gene, which encodes the enzyme that converts the prodrug INH to its active form. Through mechanistic studies, we discovered that C10 inhibits Mtb respiration, revealing a link between respiration homeostasis and INH sensitivity. Therefore, by using C10 to dissect Mtb persistence, we discovered that INH resistance is not absolute and can be reversed.
As the deadliest pathogen in the world, Mycobacterium tuberculosis (Mtb) causes infections responsible for 1.6 million deaths in 2017 (1). During infection, Mtb is exposed to an arsenal of host-derived stresses; however, it responds to stress with physiological changes that allow it to tolerate these immune stresses and persist (2). These same physiological changes result in antibiotic tolerance, in which Mtb is genetically susceptible to antibiotics but exists in a physiological state rendering it recalcitrant to therapy (3–6). As a result, long courses of antibiotic therapy are required to treat tuberculosis (TB) (7), leading to the emergence of drug-resistant mutant strains of Mtb. In 2017, out of the 10 million cases of TB, an estimated 19% of newly treated cases and 43% of previously treated cases exhibited resistance to at least one of the frontline antibiotics (1). Resistance to the frontline antibiotic isoniazid (INH) is the most common form of Mtb monoresistance and is associated with treatment failure, relapse, and progression to multidrug-resistant TB (1). Together, the problems of phenotypic tolerance and genetic resistance to antibiotics undermine current TB treatment options. There is an urgent need for new strategies that shorten the duration of treatment and target both drug-tolerant and genetically drug-resistant Mtb, which requires a better understanding of how Mtb survives exposure to immune defenses and antibiotic therapy.
Previous work has demonstrated that a number of stresses are capable of inducing the formation of drug-tolerant Mtb (8–10). The most thoroughly studied inducer of drug tolerance is hypoxia. Exposure to hypoxic conditions has pleiotropic effects on the bacteria, including replication arrest (8), induced expression of dormancy-associated genes (11, 12), shifts in Mtb lipid composition (5, 13), and global shifts in Mtb metabolism and respiration (8, 14, 15). However, it remains unclear mechanistically how these changes in physiology confer tolerance to stress and antibiotics.
To address this gap in understanding, we developed a chemical screen to identify compounds that inhibit the development of hypoxia-induced stress and drug tolerance. Through this chemical approach, we identified a compound, C10, that inhibits the development of hypoxia-induced tolerance to oxidative stress and INH. In addition to blocking tolerance, C10 was found to prevent the selection for INH-resistant mutants and to resensitize an INH-resistant mutant to INH, providing evidence that INH resistance can be reversed in Mtb.
Results
C10 Blocks Hypoxia-Induced Tolerance to Oxidative Stress and INH.
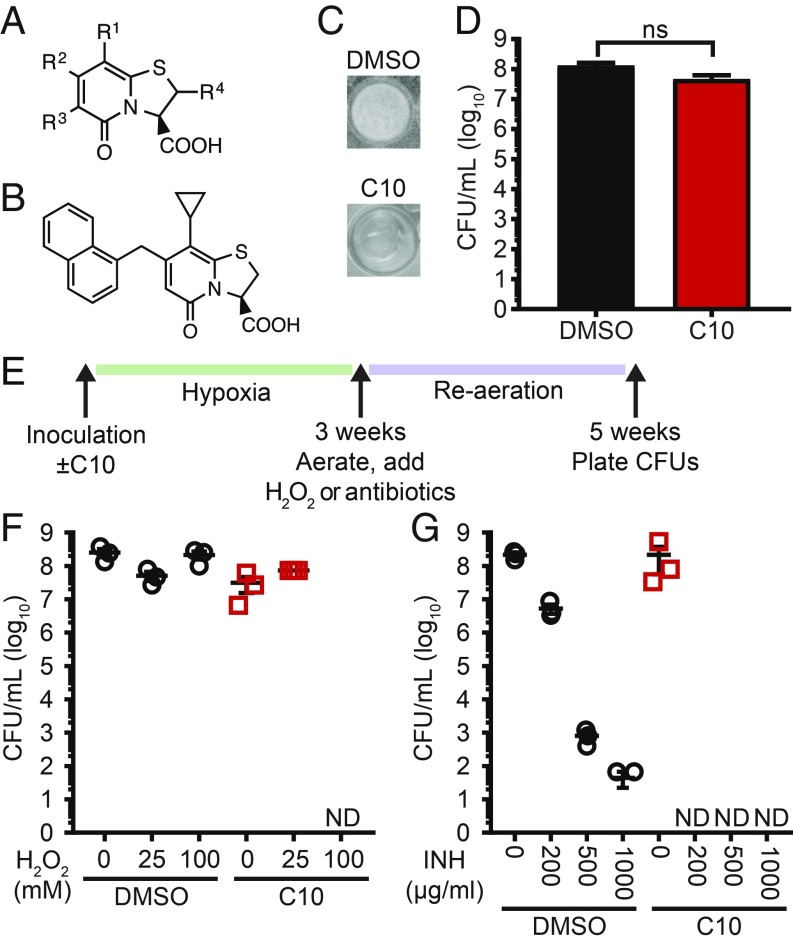
To dissect mechanisms of persistence, we used a modified version of the culture-based hypoxia model that is routinely used to study Mtb drug tolerance (8). We incubated Mtb in liquid media for 3 wk in airtight containers. During this incubation, oxygen levels dropped, and drug-tolerant bacteria developed (16). We then reaerated the cultures for an additional 2 wk, during which time Mtb formed a pellicle biofilm at the air–liquid interface. Using this model, we performed a screen for chemical inhibitors of pellicle formation. We chose a library of 91 compounds that shared a peptidomimetic bicyclic central fragment (a thiazolo ring-fused 2-pyridone; Fig. 1A). Previous work has shown that, depending on the substituents introduced to the 2-pyridone scaffold (17–20), compounds within this library exhibit diverse but highly specific biological activities, including some compounds that inhibit pellicle formation in Escherichia coli (21, 22). From this screen, we identified 12 compounds that inhibited Mtb pellicle formation at 10 μM, the most potent of which was C10 (Fig. 1B) (23). C10 inhibited Mtb pellicle formation (Fig. 1C) with a minimum inhibitory concentration of 6.25 μM (SI Appendix, Fig. S1). Despite the absence of a pellicle, wells treated with C10 contained >1 × 107 cfu/mL (Fig. 1D). Therefore, the absence of a pellicle was not due to a lack of viable bacteria; C10 specifically inhibits a physiological process required for pellicle formation.
Fig. 1.
C10 blocks hypoxia-induced tolerance to H2O2 and INH. (A) The bicyclic 2-pyridone scaffold shared by all compounds in the screening library in which compounds contained different substituents at each of the “R” groups. (B) The chemical structure of C10. (C) Mtb was incubated in low oxygen in Sauton’s medium in the presence of DMSO or 50 μM C10 for 3 wk, then reaerated and incubated for an additional 2 wk. Representative pictures from three independent experiments are shown. (D) Mtb ± 50 μM C10 was treated the same as the cultures in C, and viable cfu/mL were enumerated at 5 wk. n = 3. ns, not significant by unpaired t test. (E) Schematic of stress assays. (F and G) Mtb was cultured in low oxygen conditions ± 50 μM C10 for 3 wk, then reaerated and treated with H2O2 (F) or INH (G) for an additional 2 wk before cfu/mL were enumerated. Mean ± SEM between biological triplicates is graphed for each sample. ND, not detected; limit of detection, 67 cfu/mL. Complete statistical comparisons for all data are provided in SI Appendix, Table S1.
Because hypoxia promotes drug tolerance as well as pellicle formation (8, 16), we reasoned that small-molecule pellicle inhibitors may target physiological processes linked to stress and drug tolerance. Therefore, we first examined the effect of C10 on the sensitivity to reactive oxygen species (ROS), since mycobacteria up-regulate transcripts to cope with ROS in pellicles (24). We cultured Mtb in hypoxic conditions for 3 wk ± C10, then reaerated the cultures and added hydrogen peroxide (H2O2) to induce oxidative stress for 2 wk (Fig. 1 E and F). In the absence of C10, Mtb survived exposure to up to 100 mM H2O2 (Fig. 1F; DMSO). In contrast, exposure of C10-treated cultures to 100 mM H2O2 resulted in a cfu reduction to below the limit of detection, demonstrating that C10 blocks hypoxia-induced tolerance to H2O2 (Fig. 1F).
We then tested whether C10 affects hypoxia-induced antibiotic tolerance, starting with the frontline antibiotic INH, which targets mycolic acid biosynthesis (25). During infection, Mtb becomes phenotypically tolerant to INH, which can be reproduced in vitro by culturing Mtb in low oxygen (3, 4, 8). We incubated Mtb in hypoxic conditions for 3 wk ± C10, then reaerated the cultures and added INH for an additional 2 wk (Fig. 1 E and G). The survival of DMSO-treated control cultures decreased with increasing INH concentrations (Fig. 1G; DMSO); however, even at the highest INH concentration (1,000 μg/mL), a population of Mtb remained viable, similar to previous reports (16). The presence of C10 led to a dramatic decrease in survival following INH treatment (Fig. 1G). Exposure to 200 μg/mL INH killed only ∼1.5 logs of DMSO-treated Mtb, whereas no culturable bacteria were detected in C10-treated samples exposed to 200 μg/mL INH. Thus, C10 blocks the ability of Mtb to develop hypoxia-induced INH tolerance. In contrast, C10 did not significantly affect Mtb sensitivity to rifampicin, streptomycin, or ethambutol, which inhibit RNA polymerase, the ribosome, and arabinogalactan synthesis, respectively (26–28) (SI Appendix, Fig. S2). Therefore, C10 does not affect pan-drug tolerance induced by hypoxia and instead specifically sensitizes Mtb to INH.
C10 Potentiates Killing by INH and Prevents the Selection for INH-Resistant Mutants.
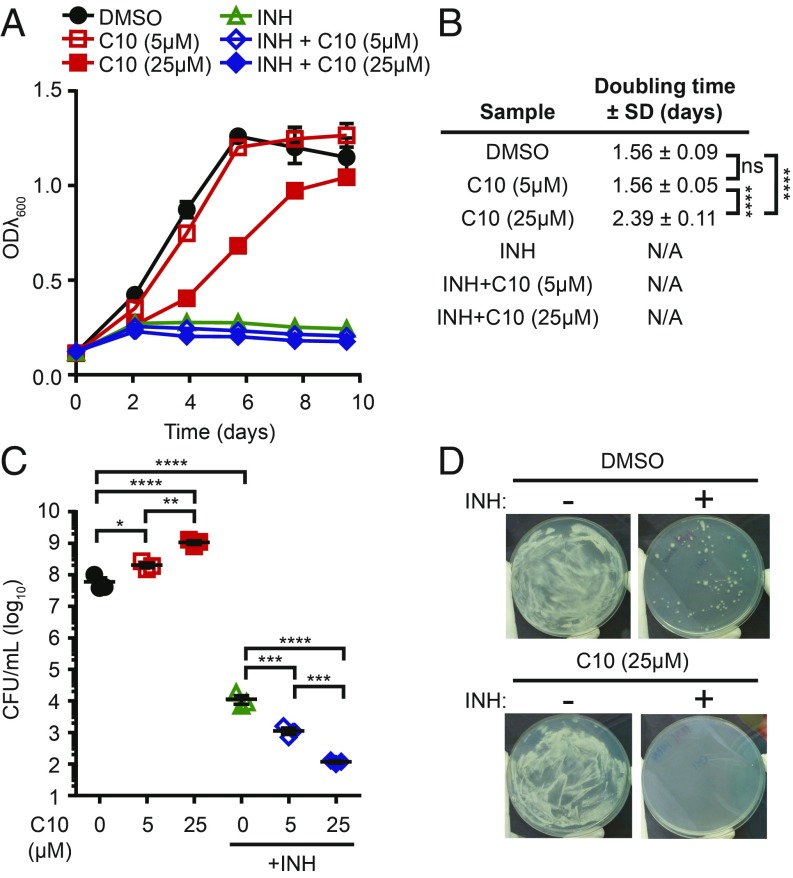
The striking and specific effects of C10 on INH tolerance indicated that C10 uniquely potentiates INH. To test whether C10 has a general effect on INH sensitivity or whether it specifically blocks hypoxia-induced INH tolerance, we cultured Mtb in planktonic, aerated conditions in media containing C10 and/or INH, and monitored growth by changes in optical density (ODλ600) (Fig. 2A). In these conditions, treatment with 5 μM C10 alone resulted in no difference in growth compared with the DMSO-treated control, and treatment with 25 μM C10 resulted in a 1.53-fold increase in Mtb doubling time (Fig. 2 A and B). Since we used an INH concentration above the minimum inhibitory concentration (0.02–0.04 μg/mL) (29), INH treatment inhibited Mtb growth with or without C10. We enumerated the surviving cfu after 10 d of treatment by plating the viable bacteria on agar media without drugs and found that the addition of C10 in combination with INH resulted in a significant and dose-dependent decrease in viable bacteria compared with INH alone. Therefore, C10 potentiates the bactericidal activity of INH against aerobically grown Mtb over a 10-d treatment period (Fig. 2C).
Fig. 2.
C10 potentiates killing by INH and prevents the selection of INH-resistant mutants. (A) WT Mtb was grown in aerated planktonic conditions in Sauton’s medium with 5 μM or 25 μM C10 ± 0.25 μg/mL INH, and ODλ600 was measured over 10 d. Mean ± SEM is graphed; n = 3. (B) The doubling time ± SD of cultures in A was calculated between day 0 and day 4. This time frame was chosen because the DMSO cultures were in the exponential growth phase. N/A indicates that growth was inhibited, and the calculation of doubling time did not accurately represent the data, as determined by R2 value (R2 <0.98). (C) After 10 d of treatment, cfu/mL were enumerated from cultures in A. Mean ± SEM values are graphed. n = 3. (D) WT Mtb was plated onto Sauton’s agar containing 0.5 μg/mL INH ± 25 μM C10. Representative pictures from three independent experiments are shown. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by one-way ANOVA with Tukey’s test. Complete statistical comparisons for all data are provided in SI Appendix, Table S1.
To further study the impact of C10 on INH efficacy, we spread ∼8 × 107 cfu of Mtb on agar media containing C10 and/or INH so that the bacteria were continually exposed to the drugs, as opposed to the transient 10-d exposure in liquid culture. Mtb formed a lawn of bacterial growth on agar containing DMSO or 25 μM C10 (Fig. 2D). Growth of Mtb on agar containing 0.5 μg/mL INH was inhibited, with the exception of spontaneous INH-resistant colonies that emerged at an approximate frequency of 1 in 106, similar to previous reports (30). In contrast, when C10 was present in combination with INH, no resistant colonies grew, demonstrating that C10 blocked the selection for INH-resistant mutants (Fig. 2D).
C10 Resensitizes katG Mutant Mtb to Inhibition by INH.
The majority of INH-resistant clinical isolates harbor mutations in katG (31), which encodes the sole catalase-peroxidase in Mtb and the enzyme that converts INH into its active form (32). We sequenced the katG gene from seven of the colonies that grew on agar containing INH (Fig. 2D) and identified katG mutations in all seven isolates; four harbored frameshift mutations, and three had missense mutations (SI Appendix, Table S2). Since no INH-resistant katG mutants grew when C10 was combined with INH (Fig. 2D), the growth of the katG mutants must be inhibited by either C10 alone or the C10-INH combination.
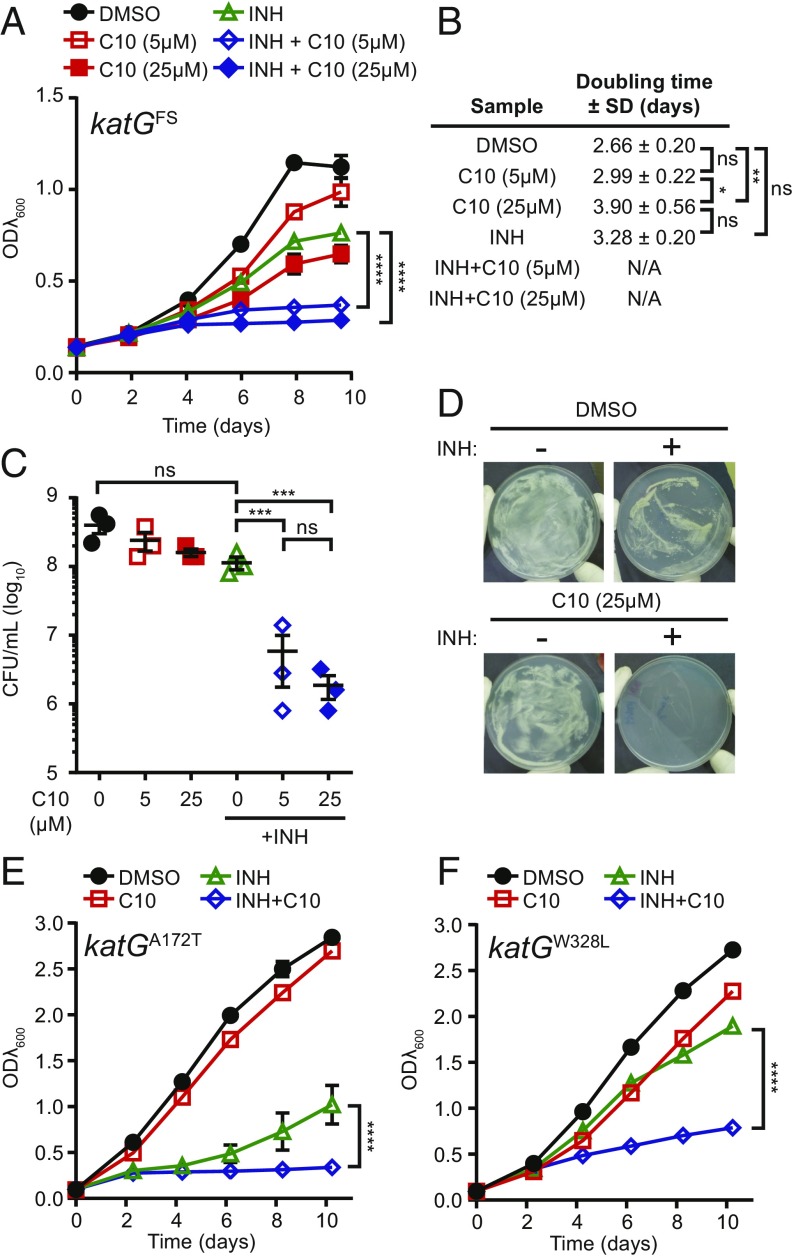
To distinguish between these possibilities, we monitored the growth of an Mtb isolate with a frameshift mutation at amino acid 6 in katG (katGFS) in aerated, planktonic cultures in the presence of C10 and/or INH (Fig. 3A). As expected, the INH-resistant katGFS mutant was able to grow in media containing INH (doubling time 3.28 ± 0.20 d), albeit at a 1.23-fold slower rate than the DMSO-treated cultures (doubling time 2.66 ± 0.20 d) (Fig. 3B). The use of 5 μM C10 did not significantly affect the growth rate of the katGFS strain, and the use of 25 μM C10 increased the doubling time of the katGFS strain by 1.47-fold (Fig. 3B), which is comparable to the 1.53-fold increase in doubling time caused by 25 μM C10 in WT Mtb (Fig. 2B). Therefore, the katGFS strain was not significantly more sensitive than WT Mtb to treatment with C10 alone. However, the combination of C10 and INH significantly inhibited growth of the katGFS strain compared with INH or C10 alone (Fig. 3A).
Fig. 3.
C10 resensitizes katG mutants to inhibition by INH. (A) katGFS Mtb was grown in Sauton’s medium with 5 μM or 25 μM C10 ± 0.25 μg/mL INH, and ODλ600 was measured over 10 d. Mean ± SEM is graphed. n = 3. (B) The doubling time of cultures in A was calculated between day 0 and day 4. This time frame was chosen to be consistent with that used in Fig. 2; however, the doubling time was similar when calculated over days 0–8. N/A indicates that growth was inhibited, and the calculation of doubling time did not accurately represent the data, as determined by R2 value (R2 <0.98). (C) After 10 d of treatment, cfu/mL were enumerated. Mean ± SEM values are graphed. n = 3. (D) katGFS Mtb was plated onto Sauton’s agar containing 0.5 μg/mL INH and/or 25 μM C10. Representative pictures from three independent experiments are shown. (E and F) Either katGA172T (E) or katGW328L (F) mutant Mtb was grown in Sauton’s medium with 5 μM C10 ± 0.25 μg/mL INH, and ODλ600 was measured over 10 d. n = 2. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant by two-way ANOVA (A, E, and F) or one-way ANOVA (B and C) with Tukey’s test. Complete statistical comparisons for all data are provided in SI Appendix, Table S1.
We enumerated viable bacteria from these cultures after 10 d of treatment by plating the surviving bacteria on agar media without drugs and found that although INH or C10 alone did not significantly decrease the number of surviving katGFS Mtb, the combination of C10 and INH resulted in a significant reduction in cfu (Fig. 3C), further demonstrating that the C10-INH combination inhibits the katGFS mutant. Similarly, the katGFS mutant grew on agar containing either C10 or INH alone, but not on agar containing the combination of both C10 and INH (Fig. 3D). Therefore, C10 restored the sensitivity of the katGFS mutant to INH.
The absence of growth of any katG mutants on plates containing INH and C10 (Fig. 2D) suggests that C10 restores the sensitivity of all katG mutants that normally would be selected for in the presence of INH alone. To directly test whether the effect of C10 can be generalized to additional INH-resistant katG mutants, we measured the impact of C10 on INH sensitivity in two additional strains harboring mutations in katG at residues that were identified as mutated in INH-resistant clinical isolates: katGA172T and katGW328L (31, 33). When we treated the katGA172T and katGW328L Mtb mutants with 5 μM C10 and/or 0.25 μg/mL INH, we found that the combination of both C10 and INH resulted in significantly decreased growth compared with either treatment alone, similar to the katGFS mutant (Fig. 3 E and F). These studies provide evidence that INH resistance can be reversed in katG mutant strains of Mtb.
C10 Perturbs Mtb Metabolism and Respiration.
To decipher the mechanism by which C10 sensitizes Mtb to INH, we compared gene expression profiles of WT Mtb treated with 5 μM or 25 μM C10 for 48 h in aerated conditions to DMSO-treated control Mtb using RNA-sequencing (RNA-seq) (SI Appendix, Table S3). Treatment with 5 μM C10 resulted in significant (Padj < 0.05) up-regulation of only 12 genes by >1.5-fold, whereas treatment with 25 μM C10 caused significant up-regulation of 194 genes by >1.5-fold, including nine of the genes up-regulated in 5 μM C10 (SI Appendix, Table S4).
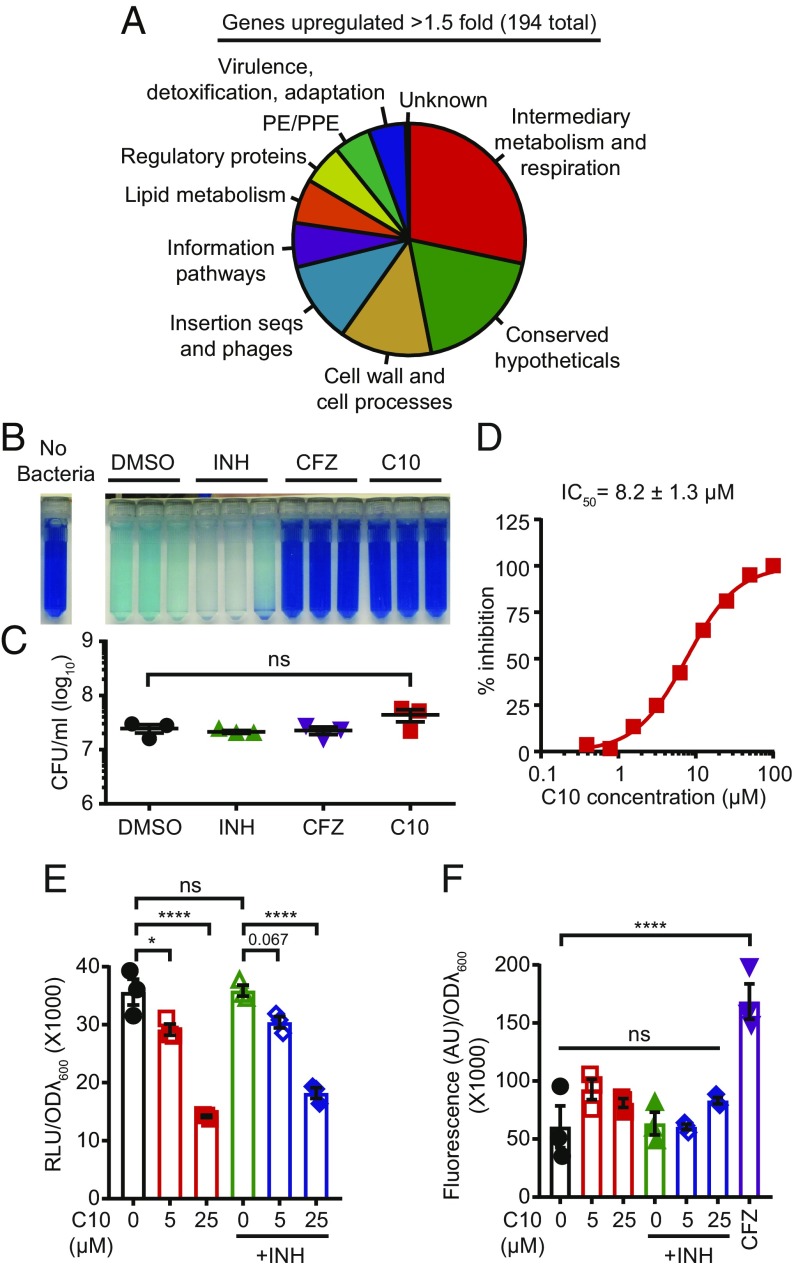
When we classified the genes induced by C10 into functional categories based on their annotation in Mycobrowser (34), we found that the functional category with the most genes up-regulated by 25 μM C10 treatment in aerobic conditions was intermediary metabolism and respiration (55 genes, Fig. 4A and SI Appendix, Table S3). In addition, the gene Rv0560c, which encodes a putative benzoquinone methyltransferase that may be involved in synthesis or modification of the electron transport chain (ETC) carrier benzoquinone, was one of the two most highly up-regulated genes in both the 5 μM and 25 μM C10 treatments. We also found that all of the genes encoding the cytochrome bd complex (cydABDC) were significantly up-regulated by >1.5-fold in Mtb treated with 25 μM C10, which is a hallmark of respiration inhibition (35). Multiple other ETC genes were also significantly induced by 25 μM C10 but did not meet the 1.5-fold cutoff (SI Appendix, Table S5).
Fig. 4.
C10 inhibits respiration in Mtb. (A) RNA-seq was performed on Mtb treated with 25 μM C10 for 48 h in aerobic conditions. The functional categories based on gene annotations in Mycobrowser for the genes significantly (Padj < 0.05) up-regulated by >1.5-fold are presented in a pie chart. (B) Mtb was pretreated for 4 h with DMSO, 2.5 μg/mL INH, 1 μg/mL CFZ, or 50 μM C10, followed by the addition of methylene blue for an additional 16 h. n = 3. (C) The viable bacteria from B were enumerated. (D) Mtb was treated with increasing concentrations of C10 and monitored for respiration in the MABA, and the IC50 ± SD was calculated with GraphPad Prism. n = 3. (E and F) WT Mtb was incubated with 5 or 25 μM C10 ± 0.25 μg/mL INH for 24 h, and either ATP levels were measured in relative luminescence units (RLU) using BacTiter Glo (E), or ROS levels were quantified by CellROX Green fluorescence in arbitrary units (AU) (F). In F, 1 μg/mL CFZ was included as a positive control. In C–F, values are mean ± SEM. *P < 0.05; ****P < 0.0001; ns, not significant by one-way ANOVA with Tukey’s test. Relevant comparisons are indicated. Complete statistics are provided in SI Appendix, Table S1.
We performed a similar gene expression analysis of Mtb cultured for 2 wk in low oxygen ± 50 μM C10 (SI Appendix, Table S6) and found that C10 caused significant (Padj < 0.05) up-regulation of 716 genes by >1.5-fold (SI Appendix, Table S7). Our finding that C10 caused up-regulation of more genes in hypoxic conditions compared with aerobic conditions may reflect the different concentrations and timing of C10 treatment in these two conditions. Alternatively, Mtb may be more sensitive to C10 treatment in hypoxic conditions, although 50 μM C10 had no effect on Mtb survival in hypoxia (Fig. 1D). Similar to the finding in aerated cultures treated with 25 μM C10, the functional category with the most up-regulated genes (148 genes) was intermediary metabolism and respiration (SI Appendix, Fig. S3).
In addition, as seen in aerobic cultures treated with 25 μM C10, we found that Rv0560c was the most highly up-regulated gene, and that multiple other ETC genes were up-regulated by >1.5-fold, including cydABDC (SI Appendix, Table S5). These data indicate that C10 affects similar pathways in both hypoxic and aerated conditions. Notably, we found that C10 did not inhibit induction of the DosR “dormancy regulon” that is up-regulated in hypoxia (12), demonstrating that C10 does not inhibit the ability of Mtb to sense or respond to hypoxia (SI Appendix, Fig. S4 and Table S8).
Since treatment with C10 in both normoxic and hypoxic conditions led to up-regulation of genes encoding components of the ETC, we examined whether C10 affects respiration by monitoring oxygen consumption by Mtb using methylene blue dye. In the presence of oxygen, this dye is blue, but when incubated with Mtb for 16 h in an airtight tube, oxygen is consumed, and the dye becomes reduced and turns colorless (36). The addition of 2.5 μg/mL INH, an antibiotic that does not target respiration, did not affect methylene blue decolorization by Mtb. In contrast, treatment with 50 μM C10 blocked methylene blue decolorization, similar to clofazimine (CFZ), which inhibits type II NADH dehydrogenases in the ETC (37) (Fig. 4B). Bacterial viability was similar in all conditions (Fig. 4C), demonstrating that inhibition of methylene blue decolorization was not a secondary effect of killing the bacteria and that C10 blocked oxygen consumption.
To quantify the minimum concentration of C10 required to perturb metabolism or respiration, we examined the activity of C10 in the microplate alamar blue assay (MABA) (38). The MABA uses the dye resazurin, which is blue in its oxidized form but is reduced to the pink fluorescent compound resorufin as a result of cellular metabolism. The MABA is commonly used to evaluate the efficacy of antimycobacterial compounds, but also serves as a measure of metabolism and respiration. C10 inhibited the reduction of resazurin with an IC50 of 8.2 ± 1.3 μM (Fig. 4D).
A limitation of the methylene blue and resazurin-based assays is that they both rely on redox-sensitive dyes as an indirect readout of metabolism and/or respiration. To confirm that C10 affects energy metabolism, we examined the effect of C10 treatment on ATP levels in the bacteria by treating Mtb with 5 μM or 25 μM C10 for 24 h and then measuring ATP levels using the luciferase-based BacTiter Glo assay (Promega). Indeed, C10 treatment caused a significant dose-responsive decrease in ATP compared with the DMSO-treated cultures (Fig. 4E). Notably, treatment with C10 in combination with INH yielded very similar results to those with C10 alone, demonstrating that INH does not enhance the ability of C10 to deplete Mtb ATP levels (Fig. 4E). As expected given the dose-responsive effects of C10 on ATP levels, increasing the concentration of C10 to 100 μM or 250 μM resulted in complete inhibition of growth (SI Appendix, Fig. S5), likely due, at least in part, to depletion of ATP.
Perturbations in respiration can result in the production of ROS (37, 39), and ROS have been shown to sensitize bacteria to INH (40–42). Therefore, if C10 treatment leads to the generation of ROS, this could contribute to increased sensitivity of Mtb to INH. To test the effect of C10 treatment on the production of ROS, we used the ROS-sensitive dye CellROX Green (Thermo Fisher Scientific), which becomes oxidized on exposure to ROS, resulting in a fluorescent product. We treated Mtb with 5 μM or 25 μM C10 for 24 h, followed by staining with CellROX Green and measuring fluorescence as a read-out for ROS. Treatment of Mtb with 1 μg/mL CFZ, a known inducer of ROS (43), increased CellROX fluorescence. In contrast, treatment with C10 did not increase CellROX fluorescence (Fig. 4F), demonstrating that C10 does not cause accumulation of ROS in WT Mtb. Adding INH to the C10-treated cultures also did not result in a change in CellROX fluorescence, indicating that the effects of C10 on INH sensitivity in WT Mtb are not mediated through ROS accumulation.
C10 Sensitizes Mtb to Acid Stress.
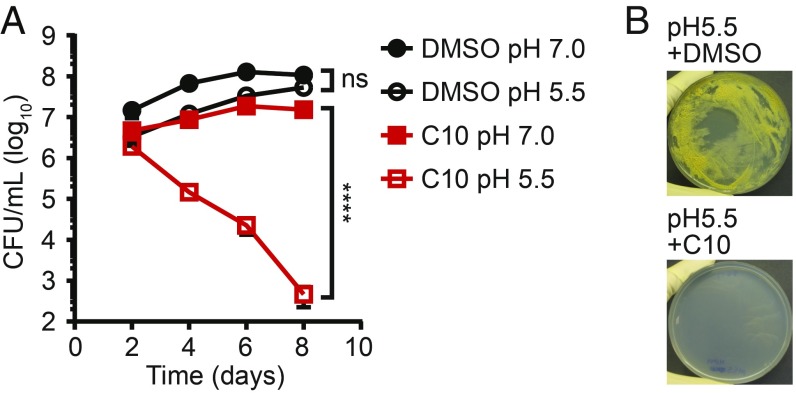
Since respiration plays an important role in maintaining intrabacterial pH homeostasis (44–46), we hypothesized that inhibition of respiration by C10 could compromise the ability of Mtb to survive exposure to acid stress. We tested whether C10 sensitizes Mtb to low pH by culturing Mtb aerobically in media at pH 7.0 or 5.5 and monitoring bacterial survival. In the absence of C10, Mtb cultured at pH 5.5 for 8 d showed no loss of viability. In contrast, in the presence of C10, the viability of Mtb cultured at pH 5.5 decreased by more than three orders of magnitude over 8 d (Fig. 5A), demonstrating that C10 sensitizes Mtb to low pH. In addition, C10 inhibited growth of Mtb on low-pH agar media (Fig. 5B), further demonstrating that C10 sensitizes Mtb to acid stress, consistent with our findings that C10 perturbs respiration.
Fig. 5.
C10 sensitizes Mtb to acid stress. (A) WT Mtb was cultured in Sauton’s media at pH 7.0 or 5.5 in the presence of DMSO or 50 μM C10, and viable bacteria were enumerated over time. n = 3. Mean ± SEM values are graphed. ****P < 0.0001; ns, not significant by two-way ANOVA with Tukey’s test. Relevant comparisons are indicated. Complete statistics are provided in SI Appendix, Table S1. (B) WT Mtb was cultured on Sauton’s agar, pH 5.5, with 25 μM C10 or DMSO, and pictures were taken at 43 d. Growth on pH 7.0 agar is shown in Fig. 2D.
C10 Potentiates Killing by the Clinical Candidate Q203 Without Targeting the Cytochrome Complexes.
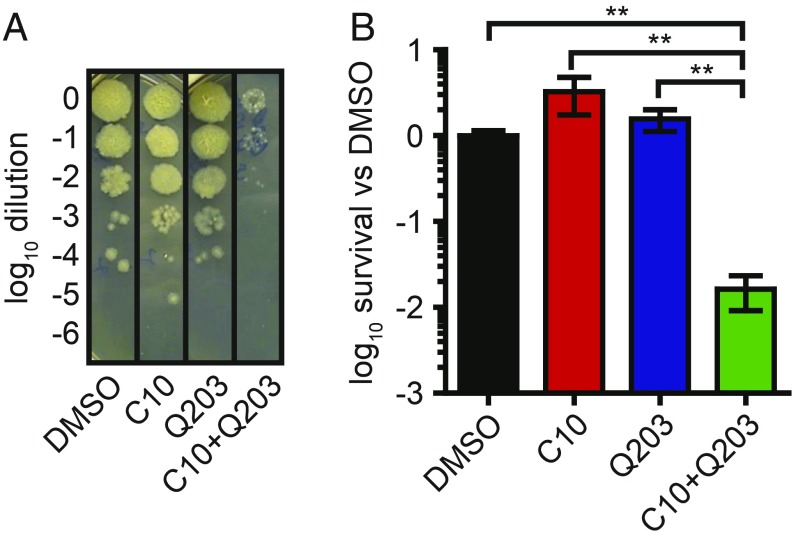
A hallmark of respiration inhibitors is their ability to synergize with genetic or chemical inhibition of parallel complexes in the ETC (43, 47). Therefore, we inquired whether C10 would enhance the activity of the ETC inhibitor Q203, which targets cytochrome bc1 and is currently in clinical trials for TB treatment (48). We incubated Mtb with 25 μM C10, 400 nM Q203, or both in liquid cultures and enumerated viable bacteria after 15 d of treatment by plating the surviving bacteria on agar media without drugs (Fig. 6 A and B). We did not observe any reduction in viable bacteria in cultures treated with C10 or Q203 alone; however, the combination of C10 and Q203 resulted in a significant decrease in Mtb viability after 15 d of treatment. These findings show that the combination of C10 with Q203 results in bactericidal activity within 15 d of treatment and support our hypothesis that C10 inhibits respiration. Mtb encodes two cytochromes, bc1 and bd, that display some redundancy in function. As such, genetic inactivation of cytochrome bd results in hypersensitivity to inhibition of cytochrome bc1 with Q203 (43, 47).
Fig. 6.
C10 potentiates killing by Q203. WT Mtb was cultured with 25 μM C10 ± 400 nM Q203 for 15 d, followed by enumeration of surviving cfu. (A) A representative image of the culture dilutions plated to enumerate cfu is shown. (B) Bacterial survival was quantified relative to DMSO-treated samples. n = 6. Values are mean ± SEM. **P < 0.01, one-way ANOVA with Tukey’s test. Relevant comparisons are indicated. Complete statistics are provided in SI Appendix, Table S1.
Since C10 induced expression of cydABDC (SI Appendix, Table S5) and also potentiated killing by Q203, we inquired whether C10 directly targeted cytochrome bd by treating a ΔcydA Mtb strain with C10 and monitoring the effects on metabolism and respiration in the MABA. We found that C10 inhibited ΔcydA Mtb to a similar extent as WT Mtb (SI Appendix, Fig. S6), demonstrating that C10 does not directly target cytochrome bd and potentiates Q203 activity via a different mechanism. These data also demonstrate that C10 does not directly target cytochrome bc1, because inhibitors of this cytochrome have increased activity in cydAB mutants (43, 47).
Discussion
The studies presented herein uniquely exploit the link between the development of drug tolerance and pellicle biofilm formation to identify compounds that inhibit pathways that contribute to drug and stress tolerance. Instead of screening for compounds that kill Mtb, we looked for inhibitors of biofilm formation and found C10, a compound that sensitizes Mtb to the antibiotics INH and Q203 as well as the physiologically relevant stresses ROS and low pH. It has also been recently shown that inhibitors of the hypoxia-responsive two-component signaling system DosRST can block hypoxia-induced INH tolerance by precluding the ability of Mtb to sense and respond to hypoxia (49). In contrast to this mechanism, C10-treated bacteria are able to sense decreases in oxygen tension and still up-regulate the DosR regulon in hypoxic conditions, indicating that C10 sensitizes Mtb to INH through a unique mechanism (SI Appendix, Fig. S4 and Table S8). Importantly, it is unlikely that the DosRST inhibitors (49) and C10 would have been identified in traditional screens for compounds that inhibit Mtb growth in traditional aerobic laboratory growth conditions. These two approaches to identify tolerance inhibitors expand on our possible TB treatment strategies.
The chemical library used in our screen included compounds that target chaperone-usher pilus biogenesis in Escherichia coli (23). However, Mtb does not encode chaperone-usher pili, indicating that this family of compounds exhibits a different mechanism of action in Mtb. In addition, C10 has been shown to inhibit the CRP transcription factor in Listeria (50, 51). Mtb encodes two CRP homologs, CRP and Cmr (52, 53). The CRP regulon (52) and the originally defined Cmr-regulated genes (53) were not universally up- or down-regulated during C10 treatment. Cmr has also been recently reported to regulate the DosR regulon (54), however, we found that the DosR regulon was appropriately down-regulated in aerated conditions and up-regulated in hypoxic conditions (SI Appendix, Tables S3, S6, and S9). Instead, our data strongly support that C10 inhibits respiration and/or metabolism in Mtb. The assays that we used cannot distinguish between the direct inhibition of respiration through inhibition of ETC enzymes and the indirect inhibition of respiration through the disruption of central carbon metabolism, which can impact respiration by affecting NADH homeostasis (55). Indeed, in addition to the up-regulation of enzymes involved in respiration, we also found up-regulation of enzymes involved in several central carbon metabolism pathways, including pyrimidine biosynthesis, propionate metabolism, and amino acid metabolism, suggesting that these pathways may be affected by C10 treatment as well (SI Appendix, Table S3).
Notably, some previous data link energy metabolism to INH sensitivity. Compounds containing free thiols that stimulate Mtb respiration enhance the bactericidal activity of INH (39); however, inhibiting respiration by interfering with menaquinone biosynthesis also enhances INH activity (35). These data indicate that perturbing respiration through either stimulation or inhibition of the ETC can result in increased INH sensitivity; however, the mechanism by which perturbations in respiration affect sensitivity to INH remains unclear. Further investigations of C10 will shed light on these phenomena.
Unlike previous studies, our present experiments with C10 demonstrate that it is possible to reverse INH resistance in an Mtb katG mutant. These findings indicate that INH resistance is not absolute and there are vulnerabilities that can be exploited to extend the clinical relevance of this antibiotic. In WT Mtb, KatG converts INH to its active form by coupling INH with NAD (32). This INH-NAD complex binds to InhA and inhibits mycolic acid synthesis, resulting in inhibition of Mtb growth (56). INH-NAD has also been shown to bind other proteins in Mtb that could be additional targets of INH (57). Our findings suggest that either (i) C10 directly or indirectly mediates the formation of the INH-NAD adduct in katG mutant strains of Mtb or (ii) C10 licenses INH to inhibit another target that does not require coupling with NAD. Distinguishing between these possibilities will be the focus of future studies and will shed light on the diversity of INH activation mechanisms and INH targets in Mtb.
In addition to our finding that resistance to a frontline TB antibiotic can be reversed, Blondiaux et al. (58) recently reported that the small molecule SMARt-420 reverses resistance to the second-line TB antibiotic ethionamide (ETH). Both INH and ETH are prodrugs that require activation in the bacteria. While the only known activator of INH is the enzyme KatG (32), ETH can be activated by multiple enzymes, including EthA and MymA, and mutations in either ethA or mymA confer a degree of resistance to ETH (59). In addition to these monooxygenases, Blondiaux et al. found that SMARt-420 led to up-regulation of another ETH-activating enzyme, EthA2. Therefore, SMARt-420 sensitizes an Mtb ethA mutant to ETH by inducing expression of this alternative ETH activation enzyme.
In addition to this example of reversing drug resistance in a genetic mutant, multiple efforts have focused on strategies to block intrinsic resistance mechanisms in Mtb. These include inhibition of β-lactamases to block the intrinsic resistance of Mtb to β-lactam antibiotics (60) and inhibition of drug efflux pump activity or cell envelope integrity to improve drug permeability and retention (61–63).
Using C10 as a chemical tool, we have uncovered a strategy to alter the physiology of Mtb so that the bacterium becomes susceptible to the stresses that it will encounter in the host as well as the frontline antibiotic INH. In particular, the unique ability of C10 to reverse INH resistance reveals that it may be possible to disarm INH resistance in the clinic, which would be of great utility to combat the global epidemic of drug-resistant TB. Future studies to identify the target of C10 and elucidate the mechanism by which C10 elicits these effects will uncover novel therapeutic targets that can be exploited for future drug development.
Materials and Methods
More detailed information is provided in SI Appendix, Materials and Methods.
Bacterial Strains and Growth Conditions.
Mtb Erdman was inoculated from a freezer stock into Middlebrook 7H9 liquid medium supplemented with 60 μL/L oleic acid, 5 g/L BSA, 2 g/L dextrose, 0.003 g/L catalase (OADC), 0.5% glycerol, and 0.05% Tween 80. Actively growing Mtb was then inoculated into Sauton’s liquid medium [0.5 g/L KH2PO4, 0.5 g/L MgSO4, 4.0 g/L l-asparagine, 6% glycerol, 0.05 g/L ferric ammonium citrate, 2.0 g/L citric acid, and 0.01% (wt/vol) ZnSO4] and used for experiments. Viable cfu were enumerated on Middlebrook 7H10 or 7H11 agar medium supplemented with OADC and 0.5% glycerol. ΔcydA Mtb was generated using specialized transduction as described in SI Appendix, Materials and Methods.
Hypoxia-Induced Pellicle Formation and Tolerance Assays.
Sauton’s medium was inoculated with stationary phase Mtb at a 1:100 dilution with and without C10. Culture vessels were closed tightly to restrict oxygen for 3 wk, at which point seals on the vessels were opened, and for biofilm assays, the cultures were incubated for another 2 wk, after which pictures were taken and/or cfu were enumerated. For tolerance assays, when hypoxic culture vessels were reaerated, H2O2 or antibiotic was pipetted into the media at the indicated concentrations. After 2 wk of exposure to the indicated stress, bacteria were harvested from each well, and cfu were enumerated.
Aerobic Liquid Media Growth Assays.
Mtb was inoculated into Sauton’s liquid medium supplemented with 0.05% Tween 80 at an ODλ600 of 0.1. Unless specified otherwise, the pH of the medium was adjusted to 7.0. Compounds were added at the indicated concentrations, and when noted, ODλ600 was monitored over time. Viable cfu were enumerated at the indicated time points by plating serial dilutions of the cultures on 7H11 agar medium plus OADC containing no antibiotics.
Agar Media Growth Assays.
For the experiments shown in Figs. 2D and 3D, ∼8 × 107 cfu were spread on the surface of plates containing agar medium and incubated at 37 °C with 5% C02 for 3 wk before pictures were taken. For the experiment in Fig. 5B, ∼2.5 × 108 cfu was spread over the surface of plates containing Sauton’s agar medium adjusted to pH 5.5.
Preparation of RNA and RNA-Sequencing.
In aerobic conditions, RNA sequencing was performed on RNA extracted from Mtb cultured aerobically in the presence of 5 μM or 25 μM C10 or DMSO for 48 h. For hypoxic cultures, Mtb was cultured in tightly sealed containers for 2 wk in the presence of 50 μM C10 or DMSO, followed by RNA extraction and sequencing.
Methylene Blue Assays.
Mtb was inoculated into Sauton’s medium containing 0.05% Tween 80 at an ODλ600 of 0.25 and incubated with the indicated concentration of INH, CFZ, or C10 for 4 h in 2-mL screwcap tubes at 37 °C in shaking conditions. Methylene blue was added to a final concentration of 0.003%, and cultures were incubated under shaking for another 16 h before photos were taken and cfu were enumerated.
MABA.
Logarithmically growing Mtb was inoculated into Sauton’s medium in 96 well plates with wells containing increasing concentrations of C10. Plates were incubated at 37 °C with 5% C02 for 1 wk, at which point resazurin was added and the plate was incubated at 37 °C with 5% C02 overnight. The production of fluorescent resorufin was measured by removing samples from the plate, mixing with formalin to kill the Mtb, and measuring the fluorescence on a Tecan M200 Pro plate reader with excitation λex = 530 nm and emission λem = 590 nm.
ATP Quantification.
Mtb was inoculated into Sauton’s medium with and without compounds at an ODλ600 of 0.1, followed by incubation in a roller apparatus for 24 h. An aliquot of the culture was then heat-inactivated at 95 °C for 20 min, and BacTiter Glo (Promega) was used to quantify ATP levels.
CellROX Assay to Measure ROS.
Mtb was inoculated into Sauton’s medium with and without compounds at an ODλ600 of 0.1 and incubated in a roller apparatus for 24 h. CellROX Green was then used to quantify ROS.
Supplementary Material
Acknowledgments
C.L.S. is supported by a Beckman Young Investigator Award from the Arnold and Mabel Beckman Foundation, an Interdisciplinary Research Initiative grant from the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital, and National Institutes of Health Grant R33 AI111696. F.A. is supported by a Göran Gustafsson Award, the Swedish Research Council, the Knut and Alice Wallenberg Foundation, and the Swedish Foundation for Strategic Research. C.L.S. and F.A. are supported by National Institutes of Health Grant R01 AI134847. K.F is supported by a pilot award from the Center for Women’s Infectious Disease Research at Washington University. G.A.H. is supported by National Science Foundation Graduate Research Fellowship DGE-1745038 and National Institute of General Medical Sciences Cell and Molecular Biology Training Grant GM007067. K.S.K. and H.T. were supported by postdoctoral stipends from the J.C. Kempe Foundation. R.L.K. is supported by a Potts Memorial Foundation postdoctoral fellowship. M.E.S. was supported through Washington University’s BioMedRAP. This project was funded in part by the National Institute of Allergy and Infectious Diseases under Grant U19AI110818 to the Broad Institute. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by National Cancer Institute (NIH) Cancer Center Support Grant P30CA91842 to Siteman Cancer Center and by Institute of Clinical and Translational Sciences (ICTS)/Clinical and Translational Science Award (CTSA) Grant UL1TR000448 from the National Center for Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH.
Footnotes
Conflict of interest statement: C.L.S., S.J.H., and F.A. have ownership interests in Quretech Bio AB, which licenses C10.
This article is a PNAS Direct Submission.
Data Deposition: The RNA-sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE129835).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818009116/-/DCSupplemental.
References
- 1.World Health Organization 2018 Global tuberculosis report. Available at https://apps.who.int/iris/bitstream/handle/10665/274453/9789241565646-eng.pdf?ua=1. Accessed October 4, 2018.
- 2.Mehta M, Rajmani RS, Singh A. Mycobacterium tuberculosis WhiB3 responds to vacuolar pH-induced changes in mycothiol redox potential to modulate phagosomal maturation and virulence. J Biol Chem. 2016;291:2888–2903. doi: 10.1074/jbc.M115.684597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain P, et al. Dual-reporter mycobacteriophages (Φ2DRMs) reveal preexisting Mycobacterium tuberculosis-persistent cells in human sputum. MBio. 2016;7:e01023-16. doi: 10.1128/mBio.01023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCune RM, Jr, Tompsett R. Fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique, I: The persistence of drug-susceptible tubercle bacilli in the tissues despite prolonged antimicrobial therapy. J Exp Med. 1956;104:737–762. doi: 10.1084/jem.104.5.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deb C, et al. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One. 2009;4:e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, et al. Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo. J Exp Med. 2016;213:809–825. doi: 10.1084/jem.20151248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.TB CARE I 2014. International Standards for Tuberculosis Care (TB CARE I, The Hague, The Netherlands), Ed. 3.
- 8.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baek S-H, Li AH, Sassetti CM. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 2011;9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gengenbacher M, Rao SPS, Pethe K, Dick T. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase, and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology. 2010;156:81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 11.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park H-D, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galagan JE, et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmermann M, et al. Dynamic exometabolome analysis reveals active metabolic pathways in non-replicating mycobacteria. Environ Microbiol. 2015;17:4802–4815. doi: 10.1111/1462-2920.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao SPS, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ojha AK, et al. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol. 2008;69:164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bengtsson C, Nelander H, Almqvist F. Asymmetric synthesis of 2,4,5-trisubstituted Δ2-thiazolines. Chemistry. 2013;19:9916–9922. doi: 10.1002/chem.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chorell E, et al. Design and synthesis of C-2 substituted thiazolo and dihydrothiazolo ring-fused 2-pyridones: Pilicides with increased antivirulence activity. J Med Chem. 2010;53:5690–5695. doi: 10.1021/jm100470t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bengtsson C, Almqvist F. Regioselective halogenations and subsequent Suzuki-Miyaura coupling onto bicyclic 2-pyridones. J Org Chem. 2010;75:972–975. doi: 10.1021/jo902458g. [DOI] [PubMed] [Google Scholar]
- 20.Chorell E, Das P, Almqvist F. Diverse functionalization of thiazolo ring-fused 2-pyridones. J Org Chem. 2007;72:4917–4924. doi: 10.1021/jo0704053. [DOI] [PubMed] [Google Scholar]
- 21.Cegelski L, et al. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson EK, et al. Modulation of curli assembly and pellicle biofilm formation by chemical and protein chaperones. Chem Biol. 2013;20:1245–1254. doi: 10.1016/j.chembiol.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emtenäs H, Åhlin K, Pinkner JS, Hultgren SJ, Almqvist F. Design and parallel solid-phase synthesis of ring-fused 2-pyridinones that target pilus biogenesis in pathogenic bacteria. J Comb Chem. 2002;4:630–639. doi: 10.1021/cc020032d. [DOI] [PubMed] [Google Scholar]
- 24.Ojha A, Hatfull GF. The role of iron in Mycobacterium smegmatis biofilm formation: The exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol Microbiol. 2007;66:468–483. doi: 10.1111/j.1365-2958.2007.05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winder FG, Collins PB. Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. J Gen Microbiol. 1970;63:41–48. doi: 10.1099/00221287-63-1-41. [DOI] [PubMed] [Google Scholar]
- 26.Hartmann G, Honikel KO, Knüsel F, Nüesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta. 1967;145:843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 27.Luzzatto L, Apirion D, Schlessinger D. Mechanism of action of streptomycin in E. coli: Interruption of the ribosome cycle at the initiation of protein synthesis. Proc Natl Acad Sci USA. 1968;60:873–880. doi: 10.1073/pnas.60.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takayama K, Armstrong EL, Kunugi KA, Kilburn JO. Inhibition by ethambutol of mycolic acid transfer into the cell wall of Mycobacterium smegmatis. Antimicrob Agents Chemother. 1979;16:240–242. doi: 10.1128/aac.16.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rastogi N, Labrousse V, Goh KS. In vitro activities of fourteen antimicrobial agents against drug susceptible and resistant clinical isolates of Mycobacterium tuberculosis and comparative intracellular activities against the virulent H37Rv strain in human macrophages. Curr Microbiol. 1996;33:167–175. doi: 10.1007/s002849900095. [DOI] [PubMed] [Google Scholar]
- 30.Bergval IL, Schuitema ARJ, Klatser PR, Anthony RM. Resistant mutants of Mycobacterium tuberculosis selected in vitro do not reflect the in vivo mechanism of isoniazid resistance. J Antimicrob Chemother. 2009;64:515–523. doi: 10.1093/jac/dkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seifert M, Catanzaro D, Catanzaro A, Rodwell TC. Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: A systematic review. PLoS One. 2015;10:e0119628. doi: 10.1371/journal.pone.0119628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei B, Wei C-J, Tu S-C. Action mechanism of antitubercular isoniazid: Activation by Mycobacterium tuberculosis KatG, isolation, and characterization of InhA inhibitor. J Biol Chem. 2000;275:2520–2526. doi: 10.1074/jbc.275.4.2520. [DOI] [PubMed] [Google Scholar]
- 33.Hazbón MH, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006;50:2640–2649. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapopoulou A, Lew JM, Cole ST. The MycoBrowser portal: A comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 2011;91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Sukheja P, et al. A novel small-molecule inhibitor of the Mycobacterium tuberculosis demethylmenaquinone methyltransferase MenG is bactericidal to both growing and nutritionally deprived persister cells. MBio. 2017;8:e02022-16. doi: 10.1128/mBio.02022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boshoff HIM, et al. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: Novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 37.Yano T, et al. Reduction of clofazimine by mycobacterial type 2 NADH:quinone oxidoreductase: A pathway for the generation of bactericidal levels of reactive oxygen species. J Biol Chem. 2011;286:10276–10287. doi: 10.1074/jbc.M110.200501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho S, Lee HS, Franzblau S. Microplate alamar blue assay (MABA) and low oxygen recovery assay (LORA) for Mycobacterium tuberculosis. In: Parish T, Roberts DM, editors. Mycobacteria Protocols. Springer; New York: 2015. pp. 281–292. [DOI] [PubMed] [Google Scholar]
- 39.Vilchèze C, et al. Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2017;114:4495–4500. doi: 10.1073/pnas.1704376114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J-Y, Burger RM, Drlica K. Role of superoxide in catalase-peroxidase-mediated isoniazid action against mycobacteria. Antimicrob Agents Chemother. 1998;42:709–711. doi: 10.1128/aac.42.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosner JL, Storz G. Effects of peroxides on susceptibilities of Escherichia coli and Mycobacterium smegmatis to isoniazid. Antimicrob Agents Chemother. 1994;38:1829–1833. doi: 10.1128/aac.38.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coulson GB, et al. Targeting Mycobacterium tuberculosis sensitivity to thiol stress at acidic pH kills the bacterium and potentiates antibiotics. Cell Chem Biol. 2017;24:993–1004.e4. doi: 10.1016/j.chembiol.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamprecht DA, et al. Turning the respiratory flexibility of Mycobacterium tuberculosis against itself. Nat Commun. 2016;7:12393. doi: 10.1038/ncomms12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan MP, et al. Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS One. 2010;5:e13356. doi: 10.1371/journal.pone.0013356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reichlen MJ, Leistikow RL, Scobey MS, Born SEM, Voskuil MI. Anaerobic Mycobacterium tuberculosis cell death stems from intracellular acidification mitigated by the DosR regulon. J Bacteriol. 2017;199:e00320-17. doi: 10.1128/JB.00320-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandrasekera NS, et al. Improved phenoxyalkylbenzimidazoles with activity against Mycobacterium tuberculosis appear to target QcrB. ACS Infect Dis. 2017;3:898–916. doi: 10.1021/acsinfecdis.7b00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalia NP, et al. Exploiting the synthetic lethality between terminal respiratory oxidases to kill Mycobacterium tuberculosis and clear host infection. Proc Natl Acad Sci USA. 2017;114:7426–7431. doi: 10.1073/pnas.1706139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pethe K, et al. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat Med. 2013;19:1157–1160. doi: 10.1038/nm.3262. [DOI] [PubMed] [Google Scholar]
- 49.Zheng H, et al. Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence. Nat Chem Biol. 2017;13:218–225. doi: 10.1038/nchembio.2259. [DOI] [PubMed] [Google Scholar]
- 50.Good JAD, et al. Attenuating Listeria monocytogenes virulence by targeting the regulatory protein PrfA. Cell Chem Biol. 2016;23:404–414. doi: 10.1016/j.chembiol.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kulén M, et al. Structure-based design of inhibitors targeting PrfA, the master virulence regulator of Listeria monocytogenes. J Med Chem. 2018;61:4165–4175. doi: 10.1021/acs.jmedchem.8b00289. [DOI] [PubMed] [Google Scholar]
- 52.Kahramanoglou C, et al. Genomic mapping of cAMP receptor protein (CRP Mt) in Mycobacterium tuberculosis: Relation to transcriptional start sites and the role of CRPMt as a transcription factor. Nucleic Acids Res. 2014;42:8320–8329. doi: 10.1093/nar/gku548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gazdik MA, Bai G, Wu Y, McDonough KA. Rv1675c (cmr) regulates intramacrophage and cyclic AMP-induced gene expression in Mycobacterium tuberculosis-complex mycobacteria. Mol Microbiol. 2009;71:434–448. doi: 10.1111/j.1365-2958.2008.06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith LJ, et al. Cmr is a redox-responsive regulator of DosR that contributes to M. tuberculosis virulence. Nucleic Acids Res. 2017;45:6600–6612. doi: 10.1093/nar/gkx406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 56.Vilchèze C, et al. Transfer of a point mutation in Mycobacterium tuberculosis InhA resolves the target of isoniazid. Nat Med. 2006;12:1027–1029. doi: 10.1038/nm1466. [DOI] [PubMed] [Google Scholar]
- 57.Argyrou A, Jin L, Siconilfi-Baez L, Angeletti RH, Blanchard JS. Proteome-wide profiling of isoniazid targets in Mycobacterium tuberculosis. Biochemistry. 2006;45:13947–13953. doi: 10.1021/bi061874m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blondiaux N, et al. Reversion of antibiotic resistance in Mycobacterium tuberculosis by spiroisoxazoline SMARt-420. Science. 2017;355:1206–1211. doi: 10.1126/science.aag1006. [DOI] [PubMed] [Google Scholar]
- 59.Grant SS, et al. Baeyer-Villiger monooxygenases EthA and MymA are required for activation of replicating and non-replicating Mycobacterium tuberculosis inhibitors. Cell Chem Biol. 2016;23:666–677. doi: 10.1016/j.chembiol.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hugonnet J-E, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abate G, et al. New verapamil analogs inhibit intracellular mycobacteria without affecting the functions of mycobacterium-specific T cells. Antimicrob Agents Chemother. 2015;60:1216–1225. doi: 10.1128/AAC.01567-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams KN, et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 2011;145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams KN, Szumowski JD, Ramakrishnan L. Verapamil, and its metabolite norverapamil, inhibit macrophage-induced, bacterial efflux pump-mediated tolerance to multiple anti-tubercular drugs. J Infect Dis. 2014;210:456–466. doi: 10.1093/infdis/jiu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.