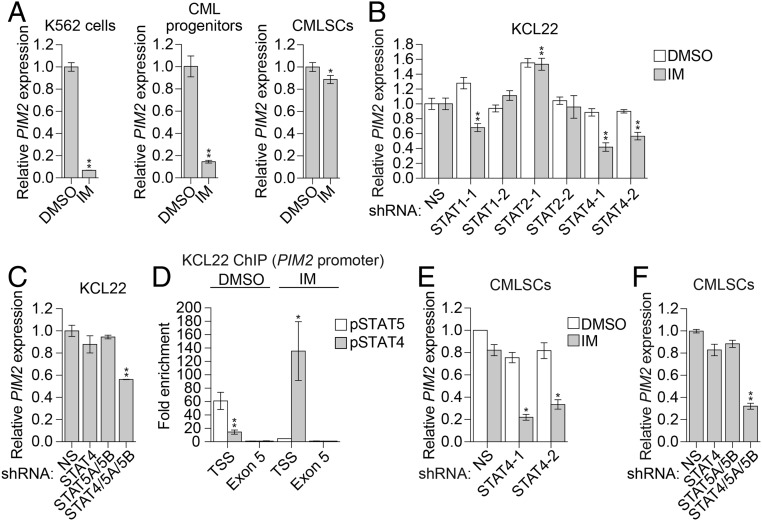
Fig. 2.
PIM2 expression in CMLSCs is promoted by both a BCR-ABL–dependent STAT5-mediated pathway and a BCR-ABL–independent STAT4-mediated pathway. (A) qRT-PCR monitoring of PIM2 expression following IM treatment in K562 cells, CML progenitors (CD34+CD38+), and CMLSCs (CD34+CD38−CD90+). Error bars indicate SD. n = 3 technical replicates of a representative experiment (out of two independent experiments). (B) qRT-PCR monitoring of PIM2 expression in KCL22 cells expressing a nonsilencing (NS) shRNA or one of two unrelated STAT1, STAT2, or STAT4 shRNAs and treated in the presence or absence of 1 µM IM. Error bars indicate SD. n = 3 technical replicates of a representative experiment (out of at least two experiments). (C) qRT-PCR monitoring of PIM2 expression in KCL22 cells expressing an NS, STAT4, and/or STAT5A/STAT5B shRNAs. Error bars indicate SD. n = 3 technical replicates of a representative experiment (out of at least two experiments). (D) ChIP analysis monitoring enrichment of phosphorylated STAT4 (pSTAT4) and phosphorylated STAT5 (pSTAT5) on the PIM2 promoter at the transcription start site (TSS) or, as a control, exon 5 in the presence or absence of IM. The results were normalized to those obtained at exon 5, which was set to 1. Error bars indicate SEM. n = 2 biological replicates. (E) qRT-PCR monitoring of PIM2 levels in CMLSCs (CD34+CD38−CD90+) expressing an NS or STAT4 shRNA and treated in the presence or absence of IM. Error bars indicate SD. n = 4 technical replicates of a single experiment. (F) qRT-PCR monitoring of PIM2 expression in CMLSCs (CD34+CD38−CD90+) expressing an NS, STAT4, and/or STAT5A/STAT5B shRNAs. Error bars indicate SD. n = 3 technical replicates of a representative experiment (out of two experiments). *P ≤ 0.05; **P ≤ 0.01.