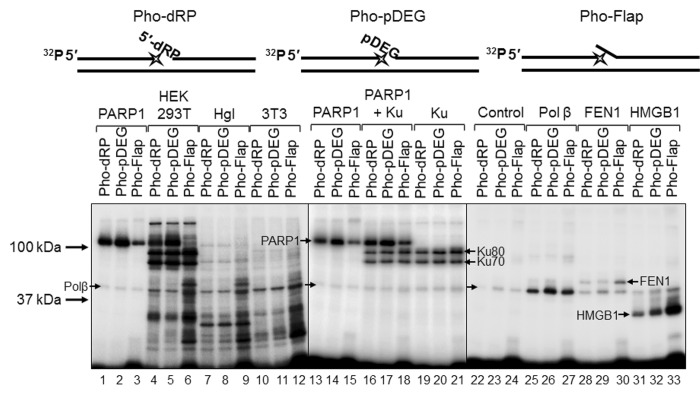
Figure 6.
Interaction of proteins with different types of photoreactive DNA. Photoaffinity modification was performed as described in the section ‘Photoaffinity modification of proteins’ using 100 nM DNAs and 1 mg/mL cell extract proteins (HEK293T, lanes 4–6; Hgl, lanes 7–9; 3T3, lanes 10–12), as well as purified PARP1 (100 nM, lanes 1–3 and 13–15), PARP1 + Ku (100 nM each, lanes 16–18), Ku (100 nM, lanes 19–21), Polβ (200 nM, lanes 25–27), FEN1 (100 nM, lanes 28–30), and HMGB1 (300 nM, lanes 31–33). Lanes 22–24 (control) correspond to the UV-light irradiated aliquots of the reaction mixtures for photoreactive DNA synthesis, which contained 100 nM Polβ. The proteins were separated by 12.5% SDS-PAGE and the proteins cross-linked to [32P]-labeled DNAs were visualized by autoradiography. The structures of the photoreactive DNAs are schematically shown at the top. The asterisk denotes the FAP-dCMP residue.