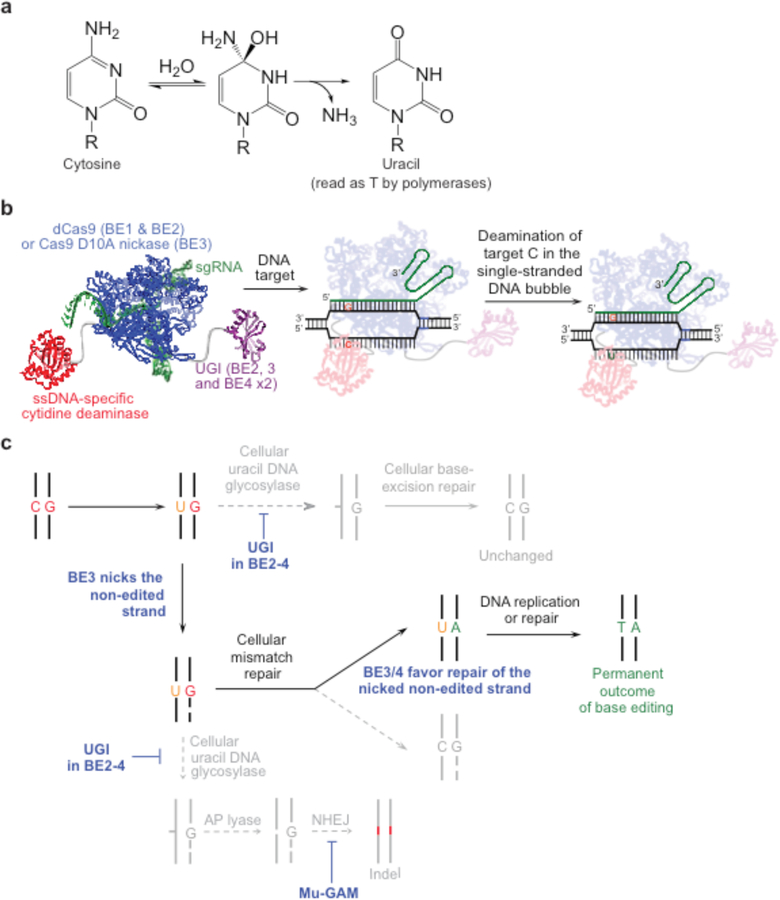
Figure 2. Cytosine base editing.
(a) Cytosine deamination generates uracil, which base pairs as thymidine. R = 2′-deoxyribose in DNA, or ribose in RNA. (b) Cytosine base editing strategy by BE1, BE2, BE3, or BE4. R-loop formation exposes a region of single-stranded DNA to the cytidine deaminase domain. Target cytosines in this region are deaminated to uracil30,39. (c) Cellular response to cytosine base editing. Uracil DNA glycosylase-mediated excision of the uracil generated in genomic DNA is inhibited by BE2, BE3, and BE4. BE3 and BE4 are designed to nick the non-edited strand (containing the G of the original C•G target base pair), stimulating cellular DNA repair of that strand to replace the G with an A, completing the conversion of the original C•G base pair to a U•A or, following DNA replication or repair to a T•A base pair30,39.