“Figure 5” (Figure to accompany Box 1).
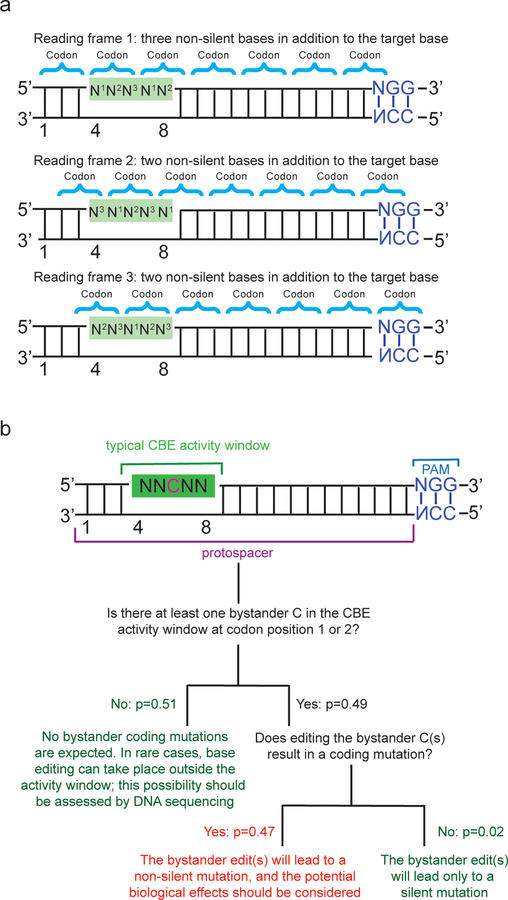
Illustration of the probability calculations for non-silent bystander editing in a protein-coding gene, as described in Box 1. (a) Use of base editing to generate a non-silent mutation in the protein-coding region of a gene. There are three possible orientations of the base editor activity window relative to the reading frame of the protein. Base editors currently generate transition mutations, and the vast majority (97%) of transitions at codon position 3 are silent. Thus, the target base in a coding gene is predominantly located at coding position 1 or 2 (N1 or N2), because the consequence of generating a transition at N3 is almost always to generate a silent mutation. In a 5-nt activity window, typical of a CBE, an average of 2.33 nucleotides at positions N1 or N2 are present in the window, in addition to the target nucleotide. For a 4-nt window typical of an ABE there are on average 1.75 nucleotides at codon positions 1 or 2 in addition to the target nucleotide (see Box 1). These additional editable nucleotides at N1 or N2, depending on their identity and sequence context, may contribute to bystander editing (see Box 1). (b) Flow chart to calculate the probability of finding a bystander mutation within a 5-nt window, as described in Box 1. The probability of the indicated event (p) assumes a genome with randomly distributed bases. For a description of how each probability was calculated, see Box 1. The same calculation is described for a 4-nt window typical of ABEs in Box 1.