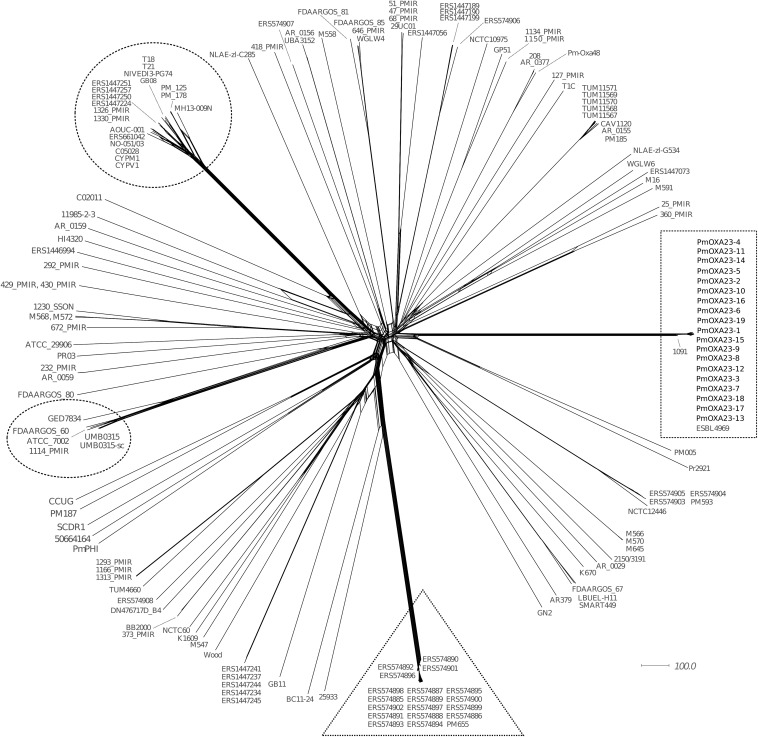
FIG 1.
Phylogenetic network of the genomes of 168 P. mirabilis isolates. The genomic comparison included the 19 blaOXA-23 isolates collected at the University Hospital of Besançon (France) and 149 P. mirabilis isolates available in the NCBI database (Table S1). The core genome was defined as genes shared by ≥95% of the selected genomes (≥160/168 genomes). It was composed of 2,660 genes of the 3,686 genes annotated in reference strain P. mirabilis HI4320. The network was built using core genome multilocus sequence typing (cgMLST) distances with the neighborNet method in SplitTree4 (17). OXA-23- and CMY-2-positive P. mirabilis clusters are surrounded by a rectangle and a triangle, respectively. Circles represent two additional genomic branches evolving from a common ancestor.