Data are lacking regarding the impact of visible pigment on rectal swab diagnostic accuracy. We describe the test characteristics of rectal swabs with and without pigment in children with gastroenteritis.
KEYWORDS: rectal swab, visible pigment, diagnostics, diarrhea, transmissible gastroenteritis virus
ABSTRACT
Data are lacking regarding the impact of visible pigment on rectal swab diagnostic accuracy. We describe the test characteristics of rectal swabs with and without pigment in children with gastroenteritis. Between December 2014 and September 2017, children (age, <18 years) with ≥3 episodes of vomiting and/or diarrhea in a 24-h period and symptoms for <7 days were enrolled through two pediatric emergency departments and from a province-wide nursing telephone advice line in Alberta, Canada. Specimens were analyzed by employing nucleic acid amplification panels. The primary outcomes were the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for the rectal swabs, with stool specimen results being used as the reference standard. An enteropathogen was detected in 76.0% (1,399/1,841) of the paired specimens. A total of 54.4% (1,001/1841) of the swabs had visible pigment. The respective enteropathogen detection characteristics of swabs with and without visible pigment were as follows: 92.2% (95% confidence interval [CI], 90.0%, 94.0%) versus 83.7% (95% CI, 80.5%, 86.4%) for sensitivity, 94.3% (95% CI, 90.5%, 96.6%) versus 91.2% (95% CI, 86.3%, 94.5%) for specificity, 97.9% (95% CI, 96.4%, 98.8%) versus 96.5% (95% CI, 94.5%, 97.8%) for PPV, and 80.9% (95% CI, 76.0%, 85.1%) versus 65.8% (95% CI, 60.0%, 71.1%) for NPV. Processing of swabs without visible pigment would increase the rate of identification of positive swabs from 50.0% (682/1,365) to 88.3% (1,205/1,365). There is a modest decrease in the reliability of a negative test on swabs without evidence of pigment, but the overall yield is significantly greater when they are not excluded from testing. Hence, rectal swabs without visible feces should not be routinely rejected from testing.
INTRODUCTION
Obtaining stool from children is challenging, particularly in outpatient settings, as a sample is frequently not provided by the child during medical visits (1), thereby prolonging the time to results because of delays in specimen acquisition (2). Handling of bulk stools also increases exposure risk. Flocked rectal swabs are a potential alternative that compares favorably with stool specimens (3–6), including when they are obtained from patients with isolated vomiting (3). Professional society guidelines endorse rectal swab use in children if timely stool specimens cannot be obtained (7), but only when visible pigment is noted on the swab (8).
Evidence to support the requirement of visible pigment is sparse. In one study, approximately 10% of rectal swab specimens received in the microbiology laboratory were excluded from processing due to the absence of visible pigment (9), and an unknown proportion were not even submitted from the point of collection for similar reasons. The absence of visible pigment on a swab is presumed to indicate that the stool sample on the swab is inadequate, but no published data support the superior sensitivity of swabs with visible feces. As such, we sought to determine if visible pigment on a swab affects the diagnostic performance of a multiplex nucleic acid test for acute gastroenteritis using rectal swabs.
MATERIALS AND METHODS
Study design.
This report is a secondary analysis of data collected as part of the Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE) study (10). From 1 December 2014 to 30 September 2017, eligible children were consecutively enrolled in pediatric emergency departments (EDs) at the Alberta Children’s Hospital (Calgary, AB, Canada) and the Stollery Children’s Hospital (Edmonton, AB, Canada). Eligible children were <18 years of age, had had at least three episodes of vomiting and/or diarrhea in the preceding 24 h, had had <7 days of symptoms, and submitted paired rectal swab and stool specimens. Bulk stool specimen results were considered the reference standard. We excluded children who had been enrolled in this study within the previous 14 days and those presenting with psychiatric concerns or neutropenia or requiring emergent medical intervention. In a second cohort, children (home participants) meeting eligibility criteria who had contacted Health Link (Alberta Health Services), a province-wide nursing triage telephone advice line, were enrolled if the recommendation provided was to continue care at home.
Informed consent was provided by caregivers; assent was obtained when participants were deemed sufficiently mature to understand study procedures and the potential benefits and harms. The research ethics boards of the University of Calgary and University of Alberta approved this study.
Specimen acquisition and processing.
A standardized data collection form was used to collect demographic and clinical information. Each participant provided a dry rectal swab (FLOQSwab; Copan Italia, Brescia, Italy) and a fecal specimen for testing. In the ED, swab specimens were obtained by a study nurse, who inserted the swab into the rectum and then rotated the swab 360°. For children enrolled through Health Link, swab and fecal specimens were collected by their caregivers using kits that were couriered to their home.
The dry rectal swab was transported in a sterile tube (Copan Italia, Brescia, Italy); fecal specimens were collected in sterile containers (catalog number V302-F; Starplex Scientific, ON, Canada). If a fecal specimen was not provided before ED discharge, caregivers were asked to collect the sample at home. Specimens were stored at room temperature for up to 12 h while awaiting retrieval by a study-funded courier, who transported the specimens to the laboratory on ice packs.
Upon specimen receipt and before processing, laboratory personnel visually inspected the swabs and scored them as having either visible fecal material or pigment (i.e., discoloration of the swab) or no indications of fecal content. After initial processing, stool samples and dry rectal swabs were stored at −80°C until nucleic acid amplification testing was performed.
Molecular testing.
Nucleic acid extracted from the rectal swabs was tested with a Luminex xTAG gastrointestinal pathogen panel (GPP; Luminex Molecular Diagnostics, ON, Canada) (6, 11). In addition, all specimens were tested using an in-house real-time PCR gastroenteritis virus panel (GVP) that detects adenovirus, astrovirus, norovirus, rotavirus, and sapovirus (12).
Nucleic acid was extracted using a NucliSENS easyMag extractor (bioMérieux, Marcy l’Etoile, France). Dry fecal swabs were immersed in 750 μl of NucliSENS easyMAG lysis buffer and mixed. Three hundred microliters of the suspension was added to Bertin SK38 soil grinding lysis bead tubes (Luminex Molecular Diagnostics) along with 10 μl of bacteriophage MS2 (Luminex Molecular Diagnostics) to a final volume of 1,000 μl. For stool specimens, nucleic acid was extracted from 100 to 150 mg of solid stool or 100 μl of liquid stool. Nucleic acid extracted from the two sample types was eluted to a final volume of 70 μl and stored at −80°C until further testing using the GPP and the GVP (12).
A 10-μl nucleic acid extract was used in the GPP assay. For the GVP assay, 5 μl of nucleic acid extract was used to generate cDNA by reverse transcription reactions with a final volume of 20 μl (12). Each duplex real-time PCR assay targeting norovirus genogroup I (GI) and GII, rotavirus and adenovirus, or sapovirus and astrovirus contained 3.5 μl of cDNA in a final volume of 10 μl. The reaction was performed using a 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA, USA). Cycle threshold (CT) values of 38 or lower were considered positive.
Outcomes.
The primary outcomes were the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for the rectal swabs with and without visible pigment for enteropathogen detection on either molecular testing platform, with stool specimen test results being used as the reference standard. Secondary outcomes included the sensitivity, specificity, PPV, and NPV for rectal swabs with and without visible pigment based on the testing platform (i.e., GVP and GPP) and pathogen targets (with combined results from GVP and GPP for viral detection). Other secondary outcomes included overall enteropathogen positivity and the CT values from the GVP platform for rectal swabs with and without visible pigment.
Statistical analysis.
For the primary outcomes, we derived 95% confidence intervals (CI) using Wilson’s procedure with a continuity correction (13, 14). We conducted subgroup analyses of the primary outcomes for (i) the presence of diarrhea or isolated vomiting at the time of specimen collection and (ii) the specimen collection group (i.e., ED versus home). Isolated vomiting was defined as vomiting without diarrhea. To evaluate if the interval between specimen collection (i.e., collection of the rectal swab earlier than collection of stool) affects specimen test characteristics, calculations were repeated with the analysis restricted to specimens collected within 24 h of each other. Because Clostridioides difficile is identified in many asymptomatic young children (15, 16), analyses were also repeated without consideration of C. difficile detection in children <2 years of age.
Test characteristics for the secondary outcomes were analyzed as described above for the primary outcomes (see Table S1 in the supplemental material). A multivariable logistic regression assessed the association between enteropathogen positivity (the dependent variable) and the presence of visible pigment (the independent variable). The model was adjusted for a priori potential confounders, including the presence of diarrhea and the collection group (i.e., ED versus home). Model coefficients were exponentiated to provide estimates of the odds ratio (OR). We compared the cycle threshold values of positive GVP tests with versus without visible pigment using the Mann-Whitney U test. Two-tailed P values of <0.05 were considered statistically significant. Analyses were conducted using SPSS (version 24.0) software (IBM Corp., Armonk, NY).
RESULTS
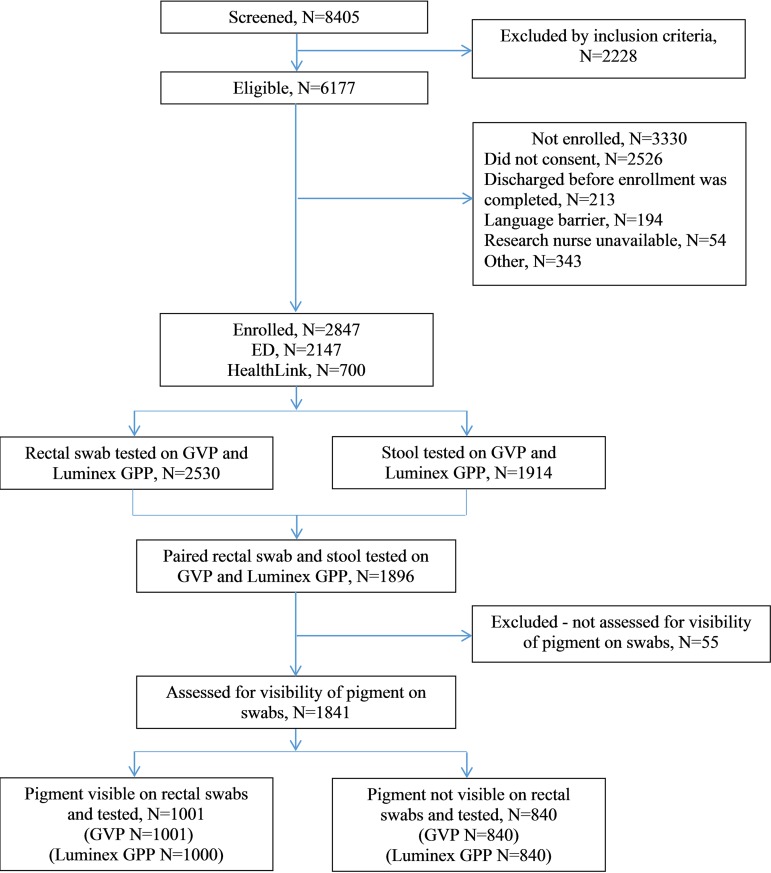
Of the 2,847 participants enrolled, 2,530 (88.8%) and 1,941 (67.2%) contributed rectal swabs and stool specimens, respectively. The 1,841 (64.7%) participants with paired specimens and documentation of the presence or absence of visible pigment were included in the analysis (Fig. 1). The participant median age was 17.8 months (interquartile range [IQR], 10.1, 36.2 months), and the median illness duration at enrollment was 46 h (IQR, 18, 89 h) (Table 1). The median time interval between illness onset and specimen acquisition was 60.3 h (IQR, 22.0, 102.5 h) for rectal swabs and 80.4 h (IQR, 42.0, 123.0 h) for stool specimens.
FIG 1.
Study participants and specimens.
TABLE 1.
Study population characteristics in relation to rectal swabs with visible pigment and rectal swabs without visible pigmentc
| Characteristic | Value for individuals providing paired stool and rectal specimens |
P valueb | ||
|---|---|---|---|---|
| All (n = 1,841) | Rectal swabs with pigment (n = 1,001) | Rectal swabs without pigment (n = 840) | ||
| Median (IQR) age (mo) | 17.8 (10.1, 36.2) | 17.3 (10.0, 36.0) | 18.4 (10.2, 37.5) | 0.27 |
| No. (%) of male patients | 998 (54.2) | 554 (55.3) | 444 (52.9) | 0.30 |
| No. (%) of patients with the following enrollment location: | ||||
| ED | 1,446 (78.5) | 731 (73.0) | 715 (85.1) | <0.001 |
| Health Link | 395 (21.5) | 270 (27.0) | ||
| No. (%) of patients with: | ||||
| Antibiotic use in past 60 days | 284 (15.4) | 159 (15.9) | 125 (14.9) | 0.60 |
| Vomiting | 1,590 (86.4) | 861 (86.0) | 729 (86.8) | 0.54 |
| Diarrhea | 1,252 (68.0) | 687 (68.6) | 565 (67.3) | 0.62 |
| Fever | 765 (41.6) | 401 (40.1) | 364 (43.3) | 0.25 |
| Median (IQR) symptom duration (h)a | 46 (18, 89) | 44 (18, 88.8) | 48 (18, 90) | 0.30 |
| Median (IQR) time (h): | ||||
| From symptom onset to rectal swab collection | 60.3 (22.0, 102.5) | 63.3 (22.1, 107.6) | 58.3 (21.1, 98.4) | 0.24 |
| From symptom onset to stool collection | 80.4 (42.0, 123.0) | 78.9 (40.1, 122.6) | 82.5 (43.6, 125.9) | 0.52 |
| Between collection of rectal swab and stool specimens | 2.1 (0.02, 21.2) | 0.5 (0, 17.5) | 7.5 (0.3, 24.8) | <0.001 |
Duration of vomiting or diarrhea, whichever was greater.
Chi-square test and Mann-Whitney tests were used for comparing categorical and continuous variables between the groups, respectively.
IQR, interquartile range; ED, emergency department.
Visible pigment was present on the rectal swab for 54.4% (1,001/1,841) of the rectal swabs with paired specimens. Rectal swabs collected at home more often had visible pigment than those submitted in the ED (home, 68.4% [270/395]; ED, 50.6% [731/1,446]; P < 0.001). There was a shorter interval between collection of rectal swabs with visible pigment and collection of the corresponding stool specimen than between collection of rectal swabs without visible pigment and collection of the corresponding stool specimen (0.5 h [IQR, 0, 17.5 h] versus 7.5 h [IQR, 0.3, 24.8 h]; P <0.001).
At least one enteropathogen was detected in 76.0% (1,399/1,841) of the paired specimens. In 160 (11.4%) pairings, only the stool specimen was positive, while in 34 (2.4%), only the rectal swab was positive (see Table S2 in the supplemental material); in 1,205 (86.1%), both were positive.
Primary outcome.
Enteropathogen detection sensitivity on swab specimens was greater when pigment was visible (92.2% [95% CI, 90.0%, 94.0%] versus 83.7% [95% CI, 80.5%, 86.4%]), and specificity was similar (94.3% [95% CI, 90.5%, 96.6%] versus 91.2% [95% CI, 86.3%, 94.5%]). Similarly, NPV was higher for swabs with visible pigment (80.9% [95% CI, 76.0% 85.1%] versus 65.8% [95% CI, 60.0%, 71.1%]) and PPV was similar (97.9% [95% CI, 96.4%, 98.8%] versus 96.5% [95% CI, 94.5%, 97.8%]) (Table 2). The finding did not change significantly when C. difficile detection in children <2 years of age was considered negative (Table S3).
TABLE 2.
Test characteristics of rectal swabs with or without visible pigment using stool specimen test results as the reference standard on either a GVP or a Luminex GPPa
| Characteristic and pigment visible on swab | No. of rectal swab specimens |
Testing characteristics of rectal swab for enteropathogen detectionb |
||||||
|---|---|---|---|---|---|---|---|---|
| Stool test positive |
Stool test negative |
|||||||
| Swab test positive | Swab test negative | Swab test positive | Swab test negative | Sensitivity (% [95% CI]) | Specificity (% [95% CI]) | PPV (% [95% CI]) | NPV (% [95% CI]) | |
| Overall | ||||||||
| Yes | 682 | 58 | 15 | 246 | 92.2 (90.0, 94.0) | 94.3 (90.5, 96.6) | 97.9 (96.4, 98.8) | 80.9 (76.0, 85.1) |
| No | 523 | 102 | 19 | 196 | 83.7 (80.5, 86.4) | 91.2 (86.3, 94.5) | 96.5 (94.5, 97.8) | 65.8 (60.0, 71.1) |
| Difference | 8.5 (4.9, 12.1) | 3.1 (−1.9, 8.5) | 1.4 (−0.6, 3.6) | 15.2 (7.9, 22.2) | ||||
| Diarrhea present at enrollment | ||||||||
| Yes | 522 | 19 | 9 | 137 | 96.5 (94.5, 97.8) | 93.8 (88.3, 96.7) | 98.3 (96.7, 99.2) | 87.8 (81.4, 92.3) |
| No | 406 | 45 | 14 | 100 | 90.0 (86.8, 92.6) | 87.7 (79.9, 92.9) | 96.7 (94.3, 98.1) | 69.0 (60.7, 76.2) |
| Difference | 6.5 (3.2, 10.0) | 6.1 (−1.5, 14.5) | 1.6 (−0.5, 4.1) | 18.9 (9.2, 28.3) | ||||
| Isolated vomiting at enrollment | ||||||||
| Yes | 160 | 39 | 6 | 108 | 80.4 (74.1, 85.5) | 94.7 (88.4, 97.8) | 96.4 (92.0, 98.5) | 73.5 (65.4, 80.3) |
| No | 115 | 57 | 5 | 95 | 66.9 (59.2, 73.7) | 95.0 (88.2, 98.1) | 95.8 (90.1, 98.5) | 62.5 (54.3, 70.1) |
| Difference | 13.5 (4.2, 22.7) | −0.3 (−7.3, 7.2) | 0.6 (−4.6, 6.7) | 11.0 (−0.09, 21.6) | ||||
| Swab collected in ED | ||||||||
| Yes | 468 | 55 | 11 | 197 | 89.5 (86.5, 91.9) | 94.7 (90.5, 97.2) | 97.7 (95.8, 98.8) | 78.2 (72.5, 83.1) |
| No | 429 | 93 | 18 | 175 | 82.2 (78.6, 85.3) | 90.7 (85.4, 94.2) | 96.0 (93.6, 97.5) | 65.3 (59.2, 70.9) |
| Difference | 7.3 (2.9, 11.7) | 4.0 (−1.5, 9.8) | 1.7 (−0.7, 4.4) | 12.9 (4.9, 20.6) | ||||
| Swab collected at home | ||||||||
| Yes | 214 | 3 | 4 | 49 | 98.6 (95.7, 99.6) | 92.5 (80.9, 97.6) | 98.2 (95.1, 99.4) | 94.2 (83.1, 98.5) |
| No | 94 | 9 | 1 | 21 | 91.3 (83.6, 95.7) | 95.5 (75.1, 99.8) | 99.0 (93.5, 99.9) | 70.0 (50.4, 84.6) |
| Difference | 7.4 (2.1, 15.1) | −3.0 (−15.3, 18.0) | −0.8 (−4.0, 4.9) | 24.2 (5.9, 44.3) | ||||
Analyses were stratified by clinical features. PPV, positive predicted value; NPV, negative predictive value; ED, emergency department.
The calculation of sensitivity, specificity, PPV, and NPV was done using test results based on stool specimens as the reference standard (see Table S3 in the supplemental material).
Sensitivity increased for both swabs with and swabs without visible pigment when children presented with diarrhea (96.5% [95% CI, 94.5%, 97.8%] versus 90.0% [95% CI, 86.8%, 92.6%]). The magnitude of the reduced sensitivity of swabs without visible pigment was greater among children with isolated vomiting (80.4% [95% CI, 74.1%, 85.5%] versus 66.9% [95% CI, 59.2%, 73.7%]). When restricted to paired specimens collected within 24 h, the findings did not significantly change (Table S4). We found a significantly reduced sensitivity of detection of C. difficile (14.5% [95% CI, 2.8, 26.2]) in swabs without visible pigment (Table 3).
TABLE 3.
Test characteristics of rectal swabs with or without visible feces using stool specimens test results as the reference standarda
| Organism and feces visibile on swab | No. of rectal swab specimens |
Testing characteristics of rectal swab for enteropathogen detectiond |
||||||
|---|---|---|---|---|---|---|---|---|
| Stool test positive |
Stool test negative |
|||||||
| Swab test positive | Swab test negative | Swab test positive | Swab test negative | Sensitivity (% [95% CI]) | Specificity (% [95% CI]) | PPV (% [95% CI]) | NPV (% [95% CI]) | |
| Adenovirus | ||||||||
| Yes | 147 | 46 | 5 | 803 | 76.2 (69.5, 82.0) | 99.4 (98.6, 99.8) | 96.7 (92.1, 98.8) | 94.6 (92.8, 96.0) |
| No | 116 | 56 | 4 | 664 | 67.4 (59.9, 74.4) | 99.4 (98.5, 99.8) | 96.7 (91.2, 98.9) | 92.2 (90.0, 94.0) |
| Difference | 8.7 (−0.9, 18.2) | -0.02 (−1.0, 1.1) | 0.04 (−4.6, 5.3) | 2.4 (−0.1, 4.9) | ||||
| Astrovirus | ||||||||
| Yes | 24 | 5 | 0 | 972 | 82.8 (64.2, 94.2) | 100 (99.6, 100) | 100 (85.8, 100) | 99.5 (98.8, 99.8) |
| No | 27 | 4 | 1 | 808 | 87.1 (70.2, 96.4) | 99.9 (99.3, 100) | 96.4 (81.7, 99.9) | 99.5 (98.7, 99.9) |
| Difference | −4.3 (−25.5, 16.5) | 0.1 (−0.4, 0.8) | 3.6 (−13.9, 20.2) | −0.02 (−0.8, 0.9) | ||||
| Norovirus GI/GII | ||||||||
| Yes | 225 | 38 | 1 | 737 | 85.6 (80.7, 89.6) | 99.9 (99.2, 100) | 99.6 (97.6, 100) | 95.1 (93.3, 96.5) |
| No | 158 | 50 | 2 | 630 | 76.0 (69.6, 81.6) | 99.7 (98.9, 100) | 98.8 (95.6, 99.8) | 92.6 (90.4, 94.5) |
| Difference | 9.6 (2.2, 17.2) | 0.2 (−0.6, 1.1) | 0.8 (−1.8, 4.5) | 2.5 (−0.1, 5.1) | ||||
| Rotavirus | ||||||||
| Yes | 184 | 11 | 4 | 802 | 94.4 (90.1, 97.2) | 99.5 (98.7, 99.9) | 97.9 (94.6, 99.4) | 98.6 (97.6, 99.3) |
| No | 150 | 20 | 4 | 666 | 88.2 (82.4, 92.7) | 99.4 (98.5, 99.8) | 97.4 (93.5, 99.3) | 97.1 (95.5, 98.2) |
| Difference | 6.1 (−0.06, 12.7) | 0.1 (−0.9, 1.2) | 0.5 (−3.5, 5.0) | 1.6 (0, 3.3) | ||||
| Sapovirus | ||||||||
| Yes | 90 | 7 | 2 | 902 | 92.8 (85.7, 97.0) | 99.8 (99.2, 100) | 97.8 (92.4, 99.7) | 99.2 (98.4, 99.7) |
| No | 60 | 14 | 4 | 762 | 81.1 (70.3, 89.3) | 99.5 (98.7, 99.9) | 93.8 (84.8, 98.3) | 98.2 (97.0, 99.0) |
| Difference | 11.7 (0.8, 23.5) | 0.3 (−0.5, 1.2) | 4.1 (−3.4, 14.0) | 1.0 (−0.1, 2.4) | ||||
| Campylobacter spp. | ||||||||
| Yes | 3 | 0 | 0 | 997 | 100 (29.2, 100) | 100 (99.6, 100) | 100 (29.2, 100) | 100 (99.6, 100) |
| No | 6 | 1 | 0 | 833 | 85.7 (42.1, 99.6) | 100 (99.6, 100) | 100 (54.1, 100) | 99.9 (99.3, 100) |
| Difference | 14.3 (−56.0, 58.0) | 0 (−0.5, 0.6) | 0 (−69.0, 48.3) | 0.1 (−0.4, 0.8) | ||||
| Clostridioides difficile | ||||||||
| Yes | 109 | 21 | 29 | 841 | 83.8 (76.4, 89.7) | 96.7 (95.2, 97.8) | 79.0 (71.2, 85.5) | 97.6 (96.3, 98.5) |
| No | 68 | 30 | 24 | 718 | 69.4 (59.3, 78.3) | 96.8 (95.2, 97.9) | 73.9 (63.7, 82.5) | 96.0 (94.3, 97.3) |
| Difference | 14.5 (2.8, 26.2) | −0.1 (−2.0, 1.8) | 5.1 (−6.4, 17.2) | 1.6 (−0.2, 3.5) | ||||
| Clostridioides difficileb | ||||||||
| Yes | 6 | 6 | 9 | 979 | 50.0 (21.1, 78.9) | 99.1 (98.3, 99.6) | 40.0 (16.3, 67.7) | 99.4 (98.7, 99.8) |
| No | 4 | 10 | 3 | 823 | 28.6 (8.4, 58.1) | 99.6 (98.9, 99.9) | 57.1 (18.4, 90.1) | 98.8 (97.8, 99.4) |
| Difference | 21.4 (−19.0, 55.0) | −0.6 (−1.5, 0.4) | −17.1 (−55.5, 28.7) | 0.6 (−0.4, 1.7) | ||||
| Escherichia coli O157:H7 | ||||||||
| Yes | 4 | 0 | 0 | 996 | 100 (39.8, 19.4) | 100 (99.6, 100) | 100 (39.8, 100) | 100 (99.6, 100) |
| No | 3 | 1 | 0 | 836 | 75.0 (19.4, 99.4) | 100 (99.6, 100) | 100 (29.2, 100) | 99.9 (99.3, 100) |
| Difference | 25.0 (−39.9, 78.1) | 0 (−0.5, 0.6) | 0 (−60.4, 69.0) | 0.1 (−0.4, 0.8) | ||||
| Salmonella spp. | ||||||||
| Yes | 17 | 3 | 1 | 979 | 85.0 (62.1, 96.8) | 99.9 (99.4, 100) | 94.4 (72.7, 99.9) | 99.7 (99.1, 99.9) |
| No | 9 | 8 | 4 | 819 | 52.9 (27.8, 77.0) | 99.5 (98.8, 99.9) | 69.2 (38.6, 90.9) | 99.0 (98.1, 99.6) |
| Difference | 32.1 (−1.2, 58.9) | 0.4 (−0.3, 1.2) | 25.2 (−6.2, 56.0) | 0.7 (−0.2, 1.7) | ||||
| Shiga toxin-producing E. coli (stx1 or stx2)c | ||||||||
| Yes | 11 | 3 | 1 | 985 | 78.6 (49.2, 95.3) | 99.9 (99.4, 100) | 91.7 (61.5, 99.9) | 99.7 (99.1, 99.9) |
| No | 6 | 3 | 1 | 830 | 66.7 (29.9, 92.5) | 99.9 (99.3, 100) | 85.7 (42.1, 99.6) | 99.6 (99.0, 99.9) |
| Difference | 11.9 (−26.5, 51.0) | 0.02 (−0.6, 0.7) | 6.0 (−28.7, 50.4) | 0.06 (−0.7, 0.9) | ||||
| Giardia | ||||||||
| Yes | 0 | 3 | 0 | 997 | 0 (0, 70.8) | 100 (99.6, 100) | NA | 99.7 (99.1, 99.9) |
| No | 1 | 2 | 0 | 837 | 33.3 (0.8, 90.6) | 100 (99.6, 100) | 100 (2.5, 100) | 99.8 (99.1, 100) |
| Difference | −33.3 (−87.5, 42.6) | 0 (−0.5, 0.6) | NA | −0.06 (−0.7, 0.7) | ||||
Analyses are stratified by individual pathogens. Pathogens found in <5 samples are not reported. NA, not available.
C. difficile-positive results for children <2.0 years of age coded as negative.
Of the 25 Shiga toxin-producing E. coli isolates (stx1 or stx2) GPP positive, 15 were culture negative; positive cultures included E. coli O157:H7 (n = 5), E. coli O145:H nonmotile (n = 1), E. coli O26:H11 (n = 1), E. coli O76:H19 (n = 1), Campylobacter jejuni (n = 1), and Shigella flexneri 2a (n = 1).
The calculation of sensitivity, specificity, PPV, and NPV was done using test results based on stool specimens as the reference standard.
Of the 1,365 participants whose stool specimen contained an enteropathogen, 682 (50.0%) of the corresponding rectal swab specimens had both visible pigment and a detected pathogen. Inclusion of rectal swabs without visible pigment but with a pathogen detected (n = 523) increased the proportion of participants with a positive stool specimen identified by use of a rectal swab to 88.3% (n = 1,205) (Table 2). Of the 34 participants whose rectal swab specimens were positive while the related bulk stool tested negative, 19 (56%) of the swabs had no visible pigment.
Secondary outcomes.
Differences in sensitivity, specificity, PPV, and NPV between swabs with and without visible pigment were consistent on both molecular diagnostic platforms (Table 4).
TABLE 4.
Test characteristics of rectal swabs with or without visible pigment using stool specimens test results as the reference standarda
| Test or organism and pigment visibile on swab | No. of rectal swab specimens |
Testing characteristics of rectal swab for enteropathogen detectionb |
||||||
|---|---|---|---|---|---|---|---|---|
| Stool test positive |
Stool test negative |
|||||||
| Swab test positive | Swab test negative | Swab test positive | Swab test negative | Sensitivity (% [95% CI]) | Specificity (% [95% CI]) | PPV (% [95% CI]) | NPV (% [95% CI]) | |
| GVP | ||||||||
| Yes | 608 | 59 | 4 | 330 | 91.2 (88.7, 93.1) | 98.8 (96.8, 99.6) | 99.4 (98.2, 99.8) | 84.8 (80.8, 88.2) |
| No | 473 | 95 | 12 | 260 | 83.3 (79.9, 86.2) | 95.6 (92.2, 97.6) | 97.5 (95.6, 98.7) | 73.2 (68.3, 77.7) |
| Difference | 7.9 (4.1, 11.8) | 3.2 (0.4, 6.7) | 1.8 (0.2, 3.8) | 11.6 (5.6, 17.6) | ||||
| Luminex GPP | ||||||||
| Yes | 527 | 54 | 25 | 394 | 90.7 (88.0, 92.9) | 94.0 (91.2, 96.0) | 95.5 (93.3, 97.0) | 88.0 (84.5, 90.8) |
| No | 412 | 84 | 20 | 324 | 83.1 (79.4, 86.2) | 94.2 (91.0, 96.3) | 95.4 (92.8, 97.1) | 79.4 (75.1, 83.2) |
| Difference | 7.6 (3.5, 11.9) | −0.2 (−3.7, 3.6) | 0.1 (−2.7, 3.1) | 8.5 (3.4, 13.7) | ||||
| Virus | ||||||||
| Yes | 609 | 58 | 6 | 328 | 91.3 (88.8, 93.3) | 98.2 (95.9, 99.3) | 99.0 (97.8, 99.6) | 85.0 (80.9, 88.3) |
| No | 474 | 96 | 12 | 258 | 83.2 (79.8, 86.1) | 95.6 (92.3, 97.6) | 97.5 (95.6, 98.7) | 72.9 (67.9, 77.4) |
| Difference | 8.2 (4.3, 12.1) | 2.7 (−0.4, 6.2) | 1.5 (−0.2, 3.5) | 12.1 (6.0, 18.1) | ||||
| Bacteria | ||||||||
| Yes | 143 | 29 | 30 | 798 | 83.1 (76.5, 88.2) | 96.4 (94.8, 97.5) | 82.7 (76.0, 87.8) | 96.5 (94.9, 97.6) |
| No | 94 | 41 | 27 | 678 | 69.6 (61.0, 77.1) | 96.2 (94.4, 97.4) | 77.7 (69.0, 84.5) | 94.3 (92.3, 95.8) |
| Difference | 13.5 (3.5, 23.5) | 0.2 (−1.8, 2.3) | 5.0 (−4.6, 15.1) | 2.2 (0.01, 4.5) | ||||
| Bacteriac | ||||||||
| Yes | 41 | 14 | 11 | 934 | 74.5 (61.0, 85.3) | 98.8 (97.9, 99.4) | 78.8 (65.3, 88.9) | 98.5 (97.5, 99.2) |
| No | 29 | 22 | 7 | 782 | 56.9 (42.2, 70.7) | 99.1 (98.2, 99.6) | 80.6 (64.0, 91.8) | 97.3 (95.9, 98.3) |
| Difference | 17.7 (−1.6, 35.5) | −0.3 (−1.4, 0.9) | −1.7 (−19.2, 17.9) | 1.3 (−0.2, 2.8) | ||||
Analyses were stratified by gastrointestinal virus panel, Luminex gastrointestinal pathogen panel, virus, and bacterium. PPV, positive predicted value; NPV, negative predictive value.
The calculation of sensitivity, specificity, PPV, and NPV was done using test results based on stool specimens as the reference standard (see Table S1 in the supplemental material).
C. difficile-positive results for children <2.0 years of age coded as negative.
Pathogen group analysis demonstrated that swabs with visible pigment had a greater sensitivity and NPV than swabs without visible pigment and specificity and PPV similar to those for swabs without visible pigment for the identification of viral pathogens (sensitivity, 91.3% [95% CI, 88.8%, 93.3%] versus 83.2% [95% CI, 79.8%, 86.1%]; NPV, 96.5% [95% CI, 94.9%, 97.6%] versus 94.3% [95% CI, 92.3%, 95.8%]) and bacterial pathogens (sensitivity, 83.1% [95% CI, 76.5%, 88.2%] versus 69.6% [95% CI, 61.0%, 77.1%]; NPV, 96.5% [95% CI, 94.9%, 97.6%] versus 94.3% [95% CI, 92.3%, 95.8%]) (Table 4). The finding did not change significantly when C. difficile detection in children <2 years of age was considered negative.
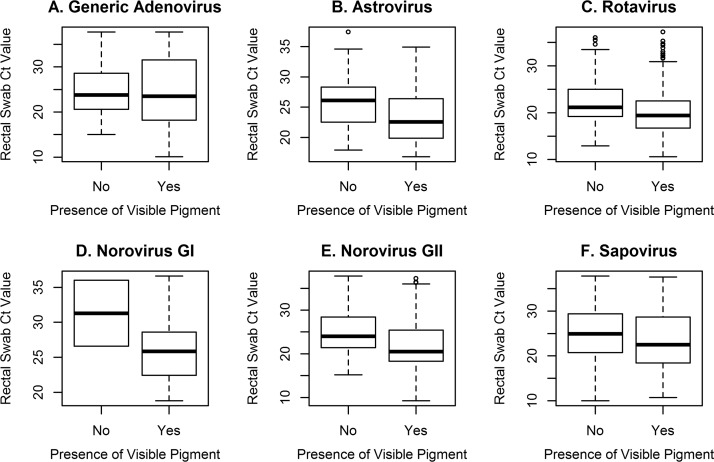
Regression analysis (n = 2,458) of overall pathogen positivity demonstrated no difference in enteropathogen detection associated with visible pigment after adjustment for diarrhea presence and sampling group (OR = 0.85; 95% CI, 0.71, 1.01; P = 0.07) (Table S5), reflecting the added diagnostic value of processing swabs without visible pigment. Lastly, rectal swabs without visible pigment had significantly higher median CT values than swabs with visible pigment on quantitative PCR testing for norovirus GII (24.0 [IQR, 21.4, 28.4] versus 20.5 [IQR, 18.3, 25.5]) and rotavirus (21.2 [IQR, 19.2, 25.1] versus 19.4 [IQR, 16.7, 22.5]) (Table S6 and Fig. 2).
FIG 2.
Box plots of real-time PCR cycle threshold (CT) values from rectal swab specimens with versus without visible pigment that were positive for a given pathogen. Generic adenovirus includes non-type 40/41.
DISCUSSION
Although swabs without visible pigment had a lower sensitivity and NPV than rectal swabs with visible pigment, when stool was employed as the reference standard, no significant difference in specificity or PPV was observed. Although swab specimens often lack visible pigment, processing them when stool is not available increased the absolute detection rate of pathogens from 50% to 88%. Although the presence of pigment is preferred, there are significant losses in diagnostic yield if swabs without pigment are rejected from testing. We suggest that laboratories consider processing flocked rectal swabs irrespective of feces visibility. Reports should note when pigment is not observed on the swab and that this may decrease the diagnostic sensitivity of the test and should prompt recollection of a specimen if it is clinically indicated.
Our findings call into question the Infectious Diseases Society of America recommendation that rectal swabs without visible feces not be tested by diagnostic microbiology laboratories (8), as we found that sensitivity was reduced by only 8.5% and the NPV was reduced by only by 15.2% in the absence of pigment. Moreover, the reduction in sensitivity was even less when children presented with diarrhea. While reduced diagnostic test characteristics are disadvantages, the overall diagnostic performance of rectal swabs without visible feces (sensitivity, 83.7% to 92.2%; specificity, 91.2% to 94.3%) is still clinically acceptable and considerably better than not testing a swab specimen at all. Rejecting swabs without visible pigment would mean that nearly 50% of rectal swabs would be discarded. While the advantages of testing a swab without visible pigment include (i) the provision of crucial and timely diagnostic information to guide therapy and public health strategy and (ii) child and care provider avoidance of the nuisance and potential risk of specimen collection at home, these benefits need to be considered in the context of the potential need to retest, which carries with it financial implications. Accepting swabs without pigment may lower the overall quality of rectal swab submissions, but we consider this unlikely for several reasons. First, the swabs analyzed for this study were collected for research purposes by nonresearch personnel (i.e., bedside nurses and caregivers); thus, there was little extrinsic motivation to ensure that there was pigment on the swab. Second, the procedure is simple and standardized. The swabs have a flange indicating the desired depth of insertion; there is little that one can do to get a better specimen. Lastly, caregiver collection at home (i.e., by untrained individuals) is as good as or better than collection by medical professionals at yielding a pathogen (3).
The lower sensitivity of rectal swabs when pigment is not visible likely reflects the reduced amount of fecal material extracted for testing and, thus, a lower pathogen load following nucleic acid extraction (12, 17). This hypothesis is supported by the higher CT values among positive rectal swabs without visible fecal material and our finding that the magnitude of the reduced detection sensitivity was greater among children with isolated vomiting. Our findings were stable across the two collection groups and the two testing platforms, suggesting that results are related to the quality of the specimen. The possibility exists that the diagnostic yield of swabs without visible feces could be increased by modifying nucleic acid extraction steps and innovations in detection technologies, as has been accomplished on buccal swabs obtained for forensic purposes (18).
A possible explanation for the overall greater reduction in the sensitivity among rectal swabs without visible pigment to detect bacteria than to detect viruses was the high rate of detection of C. difficile, which was detected in 2.8% more swabs with pigment visible than in those without visible pigment. As young children are often colonized with C. difficile (15, 19) with low pathogen loads, the C. difficile density might more often be below the detection limit in swabs without visible feces. This hypothesis is supported by evidence that among toxin-positive C. difficile-infected patients, toxin concentrations are significantly higher and CT values are significantly lower than those among carriers (20) and by our findings of the significantly reduced sensitivity of rectal swabs without pigment to detect C. difficile.
We acknowledge a number of study limitations. First, we did not assess the impact of rectal swabs without visible pigment on the sensitivity of bacterial culture. However, CT values have been demonstrated to relate to culture yield, so the results are likely transferable (21). Second, even though we assessed two molecular testing panels, alternative commercially available systems may have different sensitivity thresholds. Nonetheless, the similarity of our findings between the two panels studied suggests that our results might be generalizable to other test methods. Additionally, there were few bacterial and parasitic pathogens identified in this cohort, so more evidence regarding the use of rectal swabs without visible pigment for the detection of such pathogens and in cohorts with different pathogen distributions is needed (22). Also, a significant number of participants did not submit stool specimens and, thus, were excluded from the primary analysis; however, regression analysis included all tested rectal swab specimens, and it did not find a significant difference between the rectal swab groups.
In conclusion, if an alternate specimen is unavailable, we propose that rectal swabs without visible pigment be tested. In the event of a negative result, when clinically indicated, testing of a subsequent stool specimen or a repeat rectal swab could be considered. We suggest that laboratories comment on the visibility of pigment and test performance when reporting results from rectal swabs in order to help clinicians best interpret the results and determine if there is a need for repeat testing. Unfortunately, in our cohort there were few bacterial and parasitic pathogens; thus, additional evidence is needed to guide testing in children where such pathogens are common.
Supplementary Material
ACKNOWLEDGMENTS
We especially thank Tricia Chambers, Denise Watt, and Melissa McDougall from Health Link, Alberta Health Services, for passing the study information on to the families. We thank the patients and their families for cooperating with our study; Bryanne Crago and Christina Ferrato (Provincial Laboratory for Public Health [ProvLab]) and Judy Qiu (Department of Laboratory Medicine and Pathology, University of Alberta), DynaLIFE Dx Diagnostic Laboratory Services, community laboratories, and the Provincial Laboratory for Public Health (ProvLab), Edmonton and Calgary, especially the bacteriology staff, for their assistance with receiving, handling, and processing specimens; the emergency department research nurses and the Pediatric Emergency Medicine Research Associate Program (PEMRAP) at the Alberta Children’s Hospital for recruiting study participants; the emergency department bedside nurses for assisting with rectal swab performance; Nadia Dow and Manasi Rajagopal as well as the research assistants, research nurses, and the Little Bit of Help research volunteer program for their assistance with participant recruitment at the Stollery Children’s Hospital; the nurses at Health Link who responded to calls from across the province for their assistance with participant recruitment; and Laurel Ryan for her role as a patient adviser. We extend a special thank you to Marie Louie for building the connections that have made our endeavors possible. No compensation for the assistance of any aforementioned individuals was provided.
Flocked rectal swabs (FLOQSwab) were subsidized by Copan Italia, Brescia, Italy. Luminex Molecular Diagnostics, ON, Canada, provided support to conduct testing using the gastrointestinal pathogen panel.
This research was supported by APPETITE, which is funded by a grant from the Alberta Innovates Team Collaborative Research Innovation Opportunity. APPETITE is also supported by the Alberta Children’s Hospital Research Institute (Calgary, AB, Canada) and the Women and Children’s Health Research Institute (Edmonton, AB, Canada) through a partnership award. S.B.F. is supported by the Alberta Children’s Hospital Foundation Professorship in Child Health and Wellness. P.I.T. is also supported by grant number NIH P30DK052574 (ARAC, Digestive Diseases Research Core Center). The Pediatric Emergency Medicine Research Associate Program (PEMRAP) is supported by a grant from the Alberta Children’s Hospital Foundation.
Collaborators included Martin Lavoie, interim vice-president, population health, and chief medical health officer, Fraser Health Authority, Surrey, BC, Canada; Kimberley Simmonds, Department of Community Health Sciences, Cumming School of Medicine, University of Calgary, Calgary, AB, Canada, and Alberta Ministry of Health, Edmonton, AB, Canada; Larry Svenson, Alberta Ministry of Health, Edmonton, AB, Canada; and Nathan Zelyas, Department of Medical Microbiology and Immunology, University of Alberta, Edmonton, AB, Canada, and Provincial Laboratory for Public Health, Edmonton, AB, Canada.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00213-19.
REFERENCES
- 1.Freedman SB, Eltorki M, Chui L, Xie J, Feng S, MacDonald J, Dixon A, Ali S, Louie M, Lee BE, Osterreicher L, Thull-Freedman J. 2017. Province-wide review of pediatric Shiga toxin-producing Escherichia coli case management. J Pediatr 180:184–190.e1. doi: 10.1016/j.jpeds.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Guerrant RL, Van Gilder T, Steiner TS, Thielman NM, Slutsker L, Tauxe RV, Hennessy T, Griffin PM, DuPont H, Sack RB, Tarr P, Neill M, Nachamkin I, Reller LB, Osterholm MT, Bennish ML, Pickering LK, Infectious Diseases Society of America. 2001. Practice guidelines for the management of infectious diarrhea. Clin Infect Dis 32:331–351. doi: 10.1086/318514. [DOI] [PubMed] [Google Scholar]
- 3.Freedman SB, Xie J, Nettel-Aguirre A, Lee B, Chui L, Pang X-L, Zhuo R, Parsons B, Dickinson JA, Vanderkooi OG, Ali S, Osterreicher L, Lowerison K, Tarr PI, Chuck A, Currie G, Eltorki M, Graham T, Jiang J, Johnson D, Kellner J, Lavoie M, Louie M, MacDonald J, MacDonald S, Simmonds K, Svenson L, Tellier R, Drews S, Talbot J, Alberta Provincial Pediatric EnTeric Infection Team (APPETITE). 2017. Enteropathogen detection in children with diarrhoea, or vomiting, or both, comparing rectal flocked swabs with stool specimens: an outpatient cohort study. Lancet Gastroenterol Hepatol 2:662–669. doi: 10.1016/S2468-1253(17)30160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longtin Y, Paquet-Bolduc B, Gilca R, Garenc C, Fortin E, Longtin J, Trottier S, Gervais P, Roussy JF, Levesque S, Ben-David D, Cloutier I, Loo VG. 2016. Effect of detecting and isolating Clostridium difficile carriers at hospital admission on the incidence of C difficile infections: a quasi-experimental controlled study. JAMA Intern Med 176:796–804. doi: 10.1001/jamainternmed.2016.0177. [DOI] [PubMed] [Google Scholar]
- 5.Kabayiza JC, Andersson ME, Welinder-Olsson C, Bergstrom T, Muhirwa G, Lindh M. 2013. Comparison of rectal swabs and faeces for real-time PCR detection of enteric agents in Rwandan children with gastroenteritis. BMC Infect Dis 13:447. doi: 10.1186/1471-2334-13-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldfarb DM, Steenhoff AP, Pernica JM, Chong S, Luinstra K, Mokomane M, Mazhani L, Quaye I, Goercke I, Mahony J, Smieja M. 2014. Evaluation of anatomically designed flocked rectal swabs for molecular detection of enteric pathogens in children admitted to hospital with severe gastroenteritis in Botswana. J Clin Microbiol 52:3922–3927. doi: 10.1128/JCM.01894-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shane AL, Mody RK, Crump JA, Tarr PI, Steiner TS, Kotloff K, Langley JM, Wanke C, Warren CA, Cheng AC, Cantey J, Pickering LK. 2017. 2017 Infectious Diseases Society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 65:1963–1973. doi: 10.1093/cid/cix959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller JM, Binnicker MJ, Campbell S, Carroll KC, Chapin KC, Gilligan PH, Gonzalez MD, Jerris RC, Kehl SC, Patel R, Pritt BS, Richter SS, Robinson-Dunn B, Schwartzman JD, Snyder JW, Telford S III, Theel ES, Thomson RB Jr, Weinstein MP, Yao JD. 2018. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin Infect Dis 67:813–816. doi: 10.1093/cid/ciy584. [DOI] [PubMed] [Google Scholar]
- 9.Glisovic S, Eintracht S, Longtin Y, Oughton M, Brukner I. 2018. Rectal swab screening assays of public health importance in molecular diagnostics: sample adequacy control. J Infect Public Health 11:234–237. doi: 10.1016/j.jiph.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Freedman SB, Lee BE, Louie M, Pang XL, Ali S, Chuck A, Chui L, Currie GR, Dickinson J, Drews SJ, Eltorki M, Graham T, Jiang X, Johnson DW, Kellner J, Lavoie M, MacDonald J, MacDonald S, Svenson LW, Talbot J, Tarr P, Tellier R, Vanderkooi OG. 2015. Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE): epidemiology, emerging organisms, and economics. BMC Pediatr 15:89. doi: 10.1186/s12887-015-0407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claas EC, Burnham CA, Mazzulli T, Templeton K, Topin F. 2013. Performance of the xTAG(R) gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J Microbiol Biotechnol 23:1041–1045. doi: 10.4014/jmb.1212.12042. [DOI] [PubMed] [Google Scholar]
- 12.Pang XL, Preiksaitis JK, Lee BE. 2014. Enhanced enteric virus detection in sporadic gastroenteritis using a multi-target real-time PCR panel: a one-year study. J Med Virol 86:1594–1601. doi: 10.1002/jmv.23851. [DOI] [PubMed] [Google Scholar]
- 13.Wilson EB. 1927. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc 22:209–212. doi: 10.1080/01621459.1927.10502953. [DOI] [Google Scholar]
- 14.Newcombe RG. 1998. Interval estimation for the difference between independent proportions: comparison of eleven methods. Stat Med 17:873–890. doi:. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson MR, Van Horn GT, Tang YW, Vinje J, Payne DC, Edwards KM, Chappell JD. 2016. Using multiplex molecular testing to determine the etiology of acute gastroenteritis in children. J Pediatr 176:50–56.e2. doi: 10.1016/j.jpeds.2016.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denno DM, Shaikh N, Stapp JR, Qin X, Hutter CM, Hoffman V, Mooney JC, Wood KM, Stevens HJ, Jones R, Tarr PI, Klein EJ. 2012. Diarrhea etiology in a pediatric emergency department: a case control study. Clin Infect Dis 55:897–904. doi: 10.1093/cid/cis553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry MD, Corden SA, Howe RA. 2014. Evaluation of the Luminex xTAG gastrointestinal pathogen panel and the Savyon Diagnostics gastrointestinal infection panel for the detection of enteric pathogens in clinical samples. J Med Microbiol 63:1419–1426. doi: 10.1099/jmm.0.074773-0. [DOI] [PubMed] [Google Scholar]
- 18.Adamowicz MS, Stasulli DM, Sobestanovich EM, Bille TW. 2014. Evaluation of methods to improve the extraction and recovery of DNA from cotton swabs for forensic analysis. PLoS One 9:e116351. doi: 10.1371/journal.pone.0116351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim A, Chang JY, Shin S, Yi H, Moon JS, Ko JS, Oh S. 2017. Epidemiology and factors related to clinical severity of acute gastroenteritis in hospitalized children after the introduction of rotavirus vaccination. J Korean Med Sci 32:465–474. doi: 10.3346/jkms.2017.32.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollock NR, Banz A, Chen X, Williams D, Xu H, Cuddemi CA, Cui AX, Perrotta M, Alhassan E, Riou B, Lantz A, Miller MA, Kelly CP. 2019. Comparison of Clostridioides difficile stool toxin concentrations in adults with symptomatic infection and asymptomatic carriage using an ultrasensitive quantitative immunoassay. Clin Infect Dis 68:78–86. doi: 10.1093/cid/ciy415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wohlwend N, Tiermann S, Risch L, Risch M, Bodmer T. 2016. Evaluation of a multiplex real-time PCR assay for detecting major bacterial enteric pathogens in fecal specimens: intestinal inflammation and bacterial load are correlated in Campylobacter infections. J Clin Microbiol 54:2262–2266. doi: 10.1128/JCM.00558-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas ME. 1954. Disadvantages of the rectal swab in diagnosis of diarrhoea. Br Med J 2:394–396. doi: 10.1136/bmj.2.4884.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.