Figure 1.
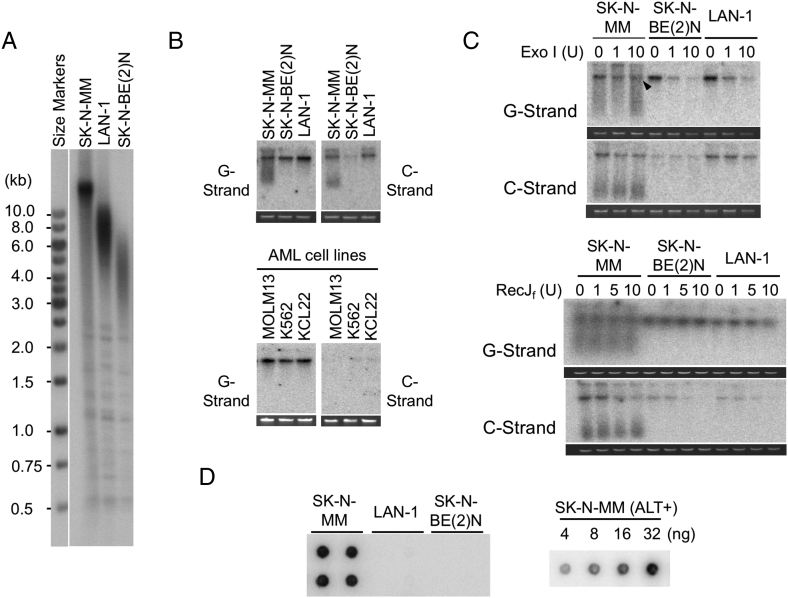
Analysis of telomere lengths and structures in neuroblastoma cell lines. A. Southern analysis for telomere restriction fragments (TRFs) was performed using DNA from one ALT positive cell line (SK-N-MM) and two telomerase positive cell lines (LAN-1 and SK-N-BE(2)N). B. Undigested genomic DNA from NB cell lines (Top) and AML cell lines (bottom) was subjected to in-gel hybridization assay to measure single-stranded DNA on the G- and C-strand. Ethidium bromide stained gels that were used for the analysis are shown below each blot. This applies to all in-gel hybridization assays presented in this paper. C. The G- and C-strand overhang signals were tested with respect to their sensitivity to Exonuclease I and RecJf. Different amounts of Exonuclease I (0 U, 1 U, and 10 U) and RecJf (0 U, 1 U, 5 U, 10 U) were used to digest 150 to 300 ng of genomic DNA at 37 °C 1 h and 2 h, respectively. Exonuclease I and RecJf degrade single-stranded 3′-overhang and 5′-overhang, respectively. Arrowhead indicates G-strand that appears to be partially resistant to Exonuclease I. D. C-circle assays were performed using 30 ng DNA from the indicated cell lines (left). All C-circle analysis of cell lines and tumor samples were performed in duplicates or quadruplets. A series of titration indicates that the assay was not saturated with up to 32 ng of SK-N-MM DNA, which exhibits the highest level of C-circles among all the analyzed samples (right).