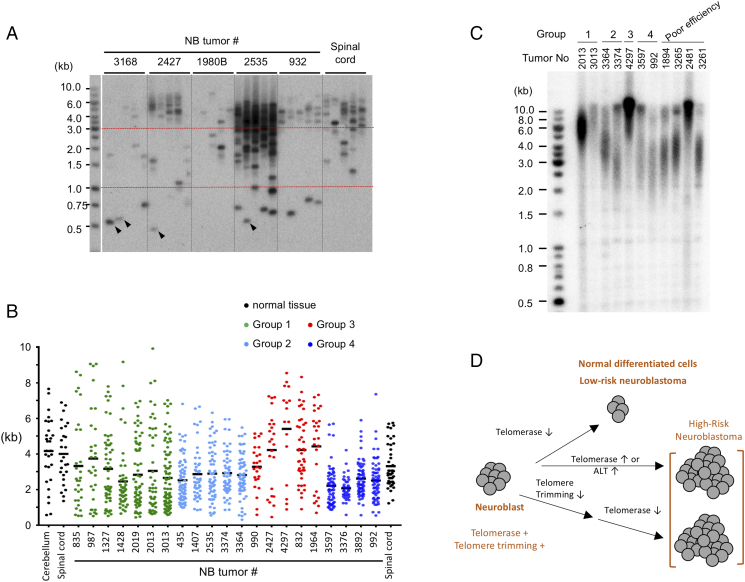
Figure 6.
STELA analysis of telomere length distributions in neuroblastoma tumors; model for the roles of telomere trimming and TMMs in NB proliferation. A. Representative STELA analysis results for the indicated NB tumors are shown. A few especially short STELA fragments that correspond to the previously described T-stumps are designated by arrowheads. B. STELA signals were analyzed by TESLA software and plotted with prism software. Based on distributions of STELA signals, NB tumors are divided into 4 groups: broad distribution (group 1), well defined telomere cluster of 1.5 to 6 kb (group 2), mostly long telomeres (>3 kb) with a minor population of short telomeres (group 3), and tightly clustered short telomeres of 1 to 3 kb (group 4). NB tumors that showed poor STELA efficiency are not included in this classification. C. Selected NB tumor DNAs were subjected to TRF Southern analysis. Ethidium bromide staining of the gel indicate that all samples have comparable amounts of DNA except for tumor 3013, which contained two- to threefold less DNA. D. Model for how telomere trimming and TMMs may influence the proliferative capacity of neuroblast-derived normal and tumor cells. The neuroblast progenitor cells are proposed to harbor both telomere trimming and telomerase activity. Normal neural tissues and low-risk neuroblastoma have limited capacity for cell division due to the retention of telomere trimming and repression of TMMs (top branch). In contrast, high-risk neuroblastoma can sustain proliferation by either activating TMMs to compensate for the telomere trimming-mediated shortening (middle branch) or by repressing telomere trimming completely prior to shutting off telomerase (middle branch).