Figure 5.
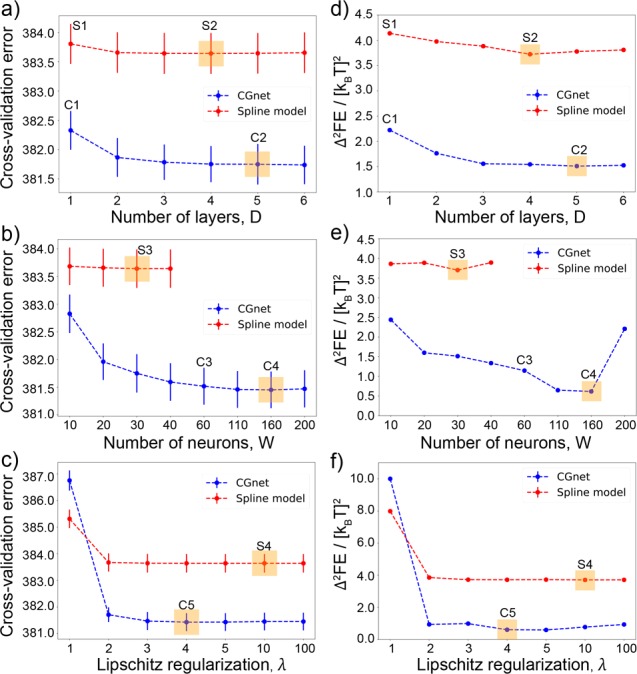
(a–c) Cross-validated force-matching error in [kcal/(mol A)]2 for the selection of the optimum structure of the network. (d–f) Difference between the two-dimensional free energy surfaces obtained from the CG models and from the reference all-atom simulations (see Figure 6) for the regularized CGnet and the spline model of alanine dipeptide. (a) Selection of the number of layers, D. (b) Selection of the number of neurons per layer, W. (c) Selection of the Lipschitz regularization strength, λ. The selected hyperparameters, corresponding to the smallest cross-validation error, are highlighted by orange boxes. Blue dashed lines represent the regularized CGnet, red dashed lines the spline model, and vertical bars the standard error of the mean. (d–f) Difference between the reference all-atom free energy surface and the free energy surfaces corresponding to the choices of hyperparameters indicated in panels a–c as (C1, C2, C3, C4, C5) for CGnet and as (S1, S2, S3, S4) for the spline model.
