Figure 4.
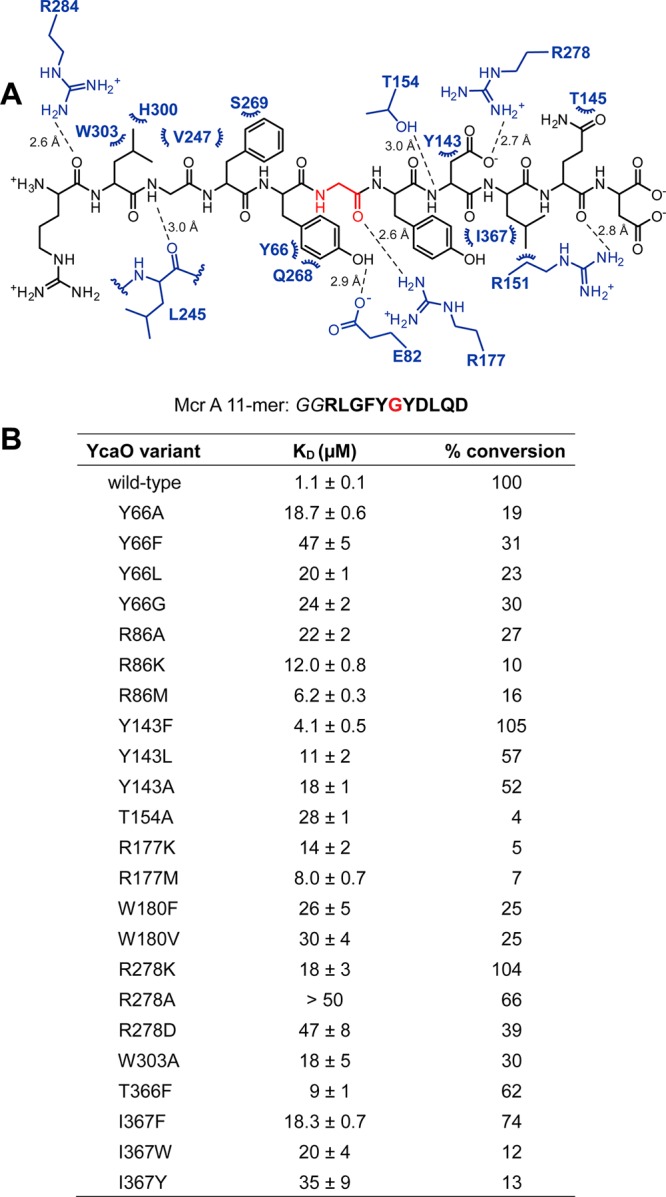
Residues important for the interactions between MjYcaO and MjMcrA 11-mer. (A) The coordinates obtained for the MjYcaO–MjMcrA 11-mer complex were processed by LigPlot Plus35 (Dimplot function) to extract the putative interactions between the protein and peptide with the default runtime parameters. Residues from the MjYcaO are in blue, and those from the MjMcrA 11-mer peptide are in black with the modified Gly highlighted in red. Hydrogen bonds are denoted by dashed lines (distances indicated next to the lines), and hydrophobic contacts are indicated by arcs. Stereochemistry is omitted for clarity. (B) Summary of the binding constants and conversion of the MjMcrA 11-mer substrate by MjYcaO variants. The synthetic peptide derived from MjMcrA [Arg(−5)–Asp(+5)] is shown with the residue naturally thioamidated Gly(0) in red. Binding constants were determined by fluorescence polarization assay with the FITC-labeled MjMcrA 11-mer peptide with Gly–Gly as a linker. Error is represented as SEM (n = 3). Conversion was measured by HPLC (n = 2) and normalized to wild-type MjYcaO.
