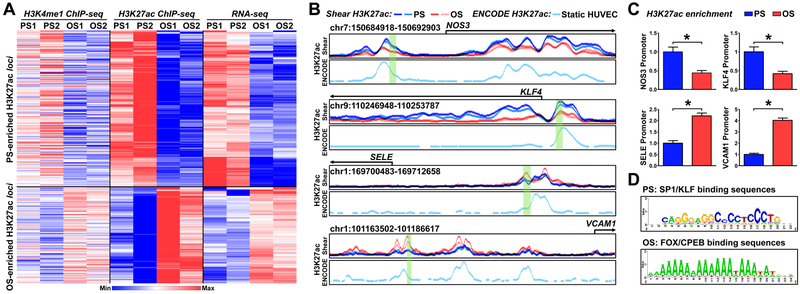
Figure 1. Shear stress regulation of histone modifications, assessed by H3K4me1 ChIP-seq, H3K27ac ChIP-seq, and RNA-seq.
(A, B) Human umbilical vein endothelial cells (HUVECs) were exposed to pulsatile flow (PS; 12 ± 4 dyn/cm2) or oscillatory flow (OS; 1 ± 4 dyn/cm2) for 16 hr. Cells from two biological repeats (PS1, OS1, PS2, OS2) were subjected to H3K4me1 ChIP-seq, and H3K27ac ChIP-seq, and RNA-seq analyses. (A) Heat map of the normalized H3K4me1 and H3K27ac enrichment and RNA-seq signal of the nearest expressed genes. The genes represented were those with H3K27ac and mRNA enrichment by PS. (B) Normalized PS (blue)- or OS (red)-associated H3K27ac ChIP signals at the promoter regions of NOS3, KLF4, SELE, and VCAM1. The H3K27ac enrichment in static HUVECs (light turquoise, below the PS and OS H3K27ac) were adopted from ENCODE (GSM733691)38. (C) HUVECs were exposed to PS or OS. H3K27ac enrichment in the region highlighted in green (in B) was evaluated by H3K27ac ChIP-qPCR. Data are mean ± SEM from 3 independent experiments. Statistical significance was determined by 2-tailed Mann-Whitney U test between PS and OS. *P < 0.05. (D) PS- or OS-enhanced H3K27ac hyper-condensed regions were analyzed by RSAT to decipher TF binding sites.