Fig. 1.
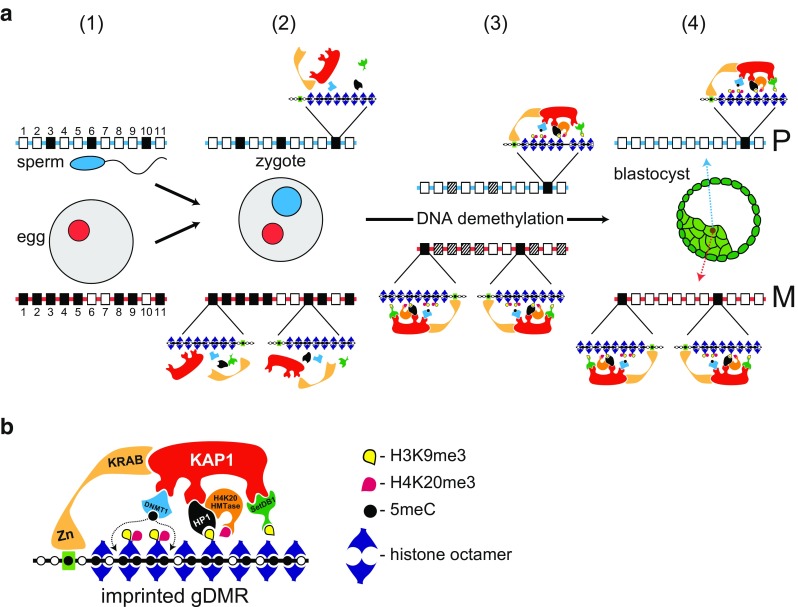
Maternal regulation of chromosomal imprinting in mice. a Preservation of methylation at imprinted gDMRs. (1) The paternal (sperm nucleus in blue) and maternal (oocyte nucleus in red) nuclei contain homologous chromosomes that carry CpG islands (CGIs) depicted as rectangles numbered 1 through to 11 on blue (paternal homolog) and red (maternal homolog) lines. Open rectangles represent non-methylated CGIs. Some methylated CGIs are shared (e.g., closed rectangle at position 3 on both parental homologs) and are not gDMRs. Some methylated CGIs are non-imprinted gDMRs (e.g., closed rectangles at position 6 on paternal chromosome and positions 2, 4, 5, 9, and 11 on the maternal chromosome) that will lose their methylation during the DNA demethylation that takes place as embryos pass through preimplantation development. A few methylated CGIs are imprinted gDMRs (closed rectangles at position 10 on the paternal chromosome and 1 and 8 on the maternal homolog) that will retain their methylation status through DNA demethylation. In actuality, there are over a thousand non-imprinted and imprinted gDMRs present in the oocyte nucleus and a few hundred will enter with the sperm (Kobayashi et al. 2012). This difference in number is reflected in the difference in closed rectangles on the maternal (red line) and paternal (blue line) homologs. (2) The maternal (in red) and paternal (in blue) pro-nuclei contain the homologous chromosomes (red and blue lines, respectively) described in (1). Of the ~ 1600 non-imprinted and imprinted gDMRs in the zygote, only a small percentage—the imprinted gDMRs—will preserve DNA methylation in the face of the DNA demethylation that takes place during pre-implantation development (Messerschmidt et al. 2014). The initial assembly of the heterochromatin-like complexes that preserve methylation at imprinted gDMRs takes place in the newly fertilized zygote (see text for details). (3) Preservation of methylation at imprinted gDMRs on the paternal (position 10) and maternal (positions 1 and 8) homologs is due to localized heterochromatin-like complexes at imprinted gDMRs. The complexes preserve DNA methylation at imprinted gDMRs throughout pre-implantation development; non-imprinted gDMRs and methylated CGIs become de-methylated (stippled rectangles). (4) Global levels of DNA methylation reach their lowest point in embryonic nuclei of the blastocyst. However, methylation at imprinted gDMRs is preserved by the heterochromatin-like complexes shown in (3), on the paternal (position 10) and maternal (position 1 and 8) homologs. P denotes paternal homolog and M the maternal homolog. b Assembly of localized heterochromatin-like complex at imprinted gDMRs. Methylation of cytosines in CpG dinucleotides (black circles) is preserved by the assembly of a heterochromatin-like complex at imprinted gDMRs. The complex is targeted by the KRAB zinc-finger protein Zfp57 that binds the hexamer motif TGCCGC when the cytosine in the CpG is methylated (black circle in green rectangle). This in turn recruits KAP1, which is a modular protein that acts as a focal point for the recruitment of Setdb1 histone methyltransferase, HP1, and Dnmt1. HP1 binds the H3K9me3 generated by Setdb1 and recruits a H4K20me3 histone methyl-transferase that generates H4K20me3 thus forming the H3K9me3:HP1:H4K20me3 pathway. DNA methylation at the imprinted gDMR is maintained (dotted lines) by Dnmt1. For a full description of the molecular constituents of the heterochromatin-like complex, see Singh (2016)
