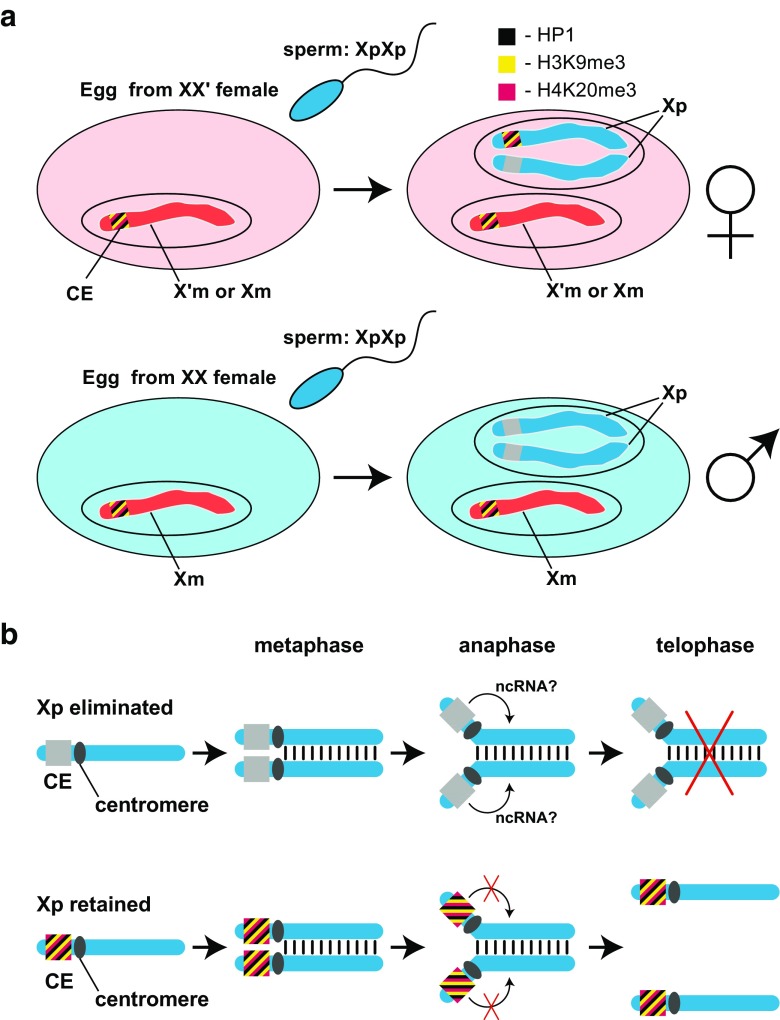
Fig. 3.
Maternal regulation of chromosomal imprinting in Sciara coprophila.a Imprinting of a paternal CE in the XX′ maternal ooplasm. In Sciara, two types of egg are fertilized by double-X (XpXp) males (sperm nuclei are given in blue). The first type are eggs laid by XX′ females (top row on left; shaded pink) that will give rise to daughters of the genetic constitution X′mXp or XmXp because eggs conditioned by XX′ mothers eliminate one Xp in the soma. The second type are eggs laid by XX females (second row on left; shaded blue) that will give rise to XmO males because eggs conditioned by XX mothers eliminate both Xps in the soma. The Xm chromosomes are not eliminated. The controlling element (CE) on the X chromosome that is embedded within heterochromomere II adjacent to the X centromere regulates the elimination of Xps. In the top row, a model is presented where the haploid maternal nucleus laid by the XX′ mother contains either an X or X′ chromosome. The maternal CE is rendered inert by the H3K9me3:HP1:H4K20me3 pathway that assembles a heterochromatin-like complex. After the fertilization by the double-X (XpXp) sperm, the paternal pro-nucleus is formed and conditioning of the ooplasm by the XX′ mother results in assembly of heterochromatin-like complex at one of the two paternal CEs rendering it inexpressible, like the maternal CE; the complex is assembled at the CE on the Xp that will later be retained (see Fig. 3b). The remaining paternal CE is an “open” expressible state. The bottom row depicts an egg laid by the XX mother. The maternal CE is again inexpressible due to the assembly of a heterochromatin-like complex, while conditioning of the ooplasm by the XX mother leaves both paternal CEs in an “open” expressible state. The stripes of color at the CE represent H3K9me3 (yellow), H4K20me3 (purple), and HP1 protein (black). Female chromosomes are in red and male are in blue. b A model for the elimination of Xp chromosomes in the embryonic soma. In the top row, the paternal CE on the Xp chromosome that will be eliminated is in an “open” expressible state. This remains so after replication where the sister chromatids become aligned and connected on the metaphase plate before separation to the poles at anaphase. At anaphase, the centromeres separate, but since the CE is active, we suggest that the CE encodes an ncRNA that affects that ability of the sister chromatids to separate. As a consequence, chromatids remain physically bound together on the metaphase plate and are eliminated. The bottom row depicts the situation in embryos that develop from eggs laid by XX′ mothers where one Xp is retained. When an Xp chromosome is retained, the putative ncRNA is not expressed because of the heterochromatin-like complex assembled at the CE. Mitosis then proceeds in the orthodox manner. The chromatids align at the metaphase plate and, first, the centromeres and then the chromosome arms separate. Each chromatid then segregates into one of the daughter nuclei. The model is taken and modified from Singh (2016)