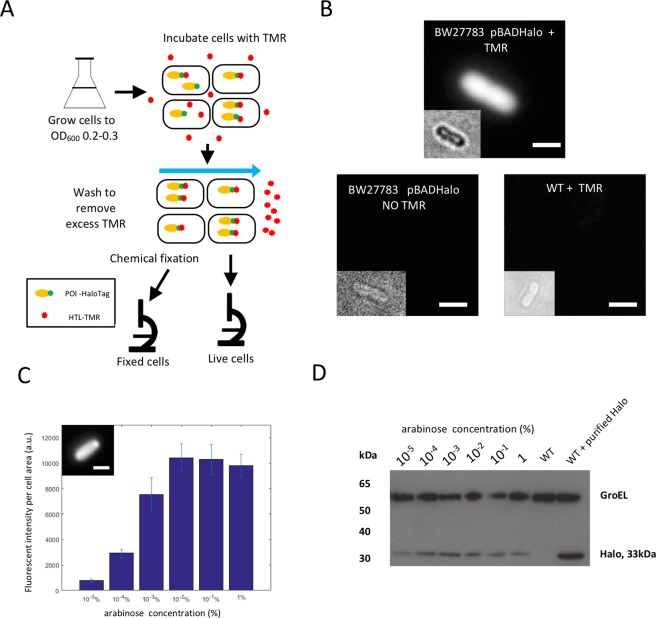
Figure 1.
Labelling of the HaloTag is specific and allows quantitative detection of protein levels. (A) Outline of the protocol used in this study. Bacterial cells were grown overnight, freshly diluted and grown until an O.D600 of approximately 0.2–0.3. They were then incubated in HTL-TMR for one hour followed by several washes. Live cells were then either immediately visualized by microscopy or chemically fixed before microscopy. (B) Specific detection of labelled HaloTag molecules in live cells. A strain expressing the HaloTag protein (arabinose concentration 1%) was incubated with HTL-TMR and showed an intense fluorescent signal (top). Virtually no signal was detected in a strain expressing the HaloTag protein not incubated with HTL-TMR (bottom left) and or in a wild-type strain incubated with HTL-TMR (bottom right). All the images are displayed using the same minimum and maximum intensity values. Insets: corresponding bright-field images defocused to identify cell contour. Bar = 1 μm. (C) The HaloTag protein expression from an arabinose inducible promoter is proportional to the arabinose concentration. The fluorescence per cell area was plotted as a function of arabinose concentration. Cells were chemically fixed immediately after washing to stop further expression of the HaloTag protein. Data correspond to the mean of at least two experiments and error bars correspond to the standard deviation of the mean. Inset: example of fixed cell expressing Halo using 1% arabinose in the growth medium. Bar 1 μm. (D) Western blot detection of HaloTag expression in the same induction conditions. Total cell lysates where prepared and probed with an antibody specific for HaloTag (Promega). This analysis showed increased HaloTag expression with increased arabinose concentration in good agreement with the quantification of the microscopy data. The GroEL is shown below to indicate that each lane has been loaded with a similar amount of protein. The full-length Western blot is presented in Supplementary Fig. 14.