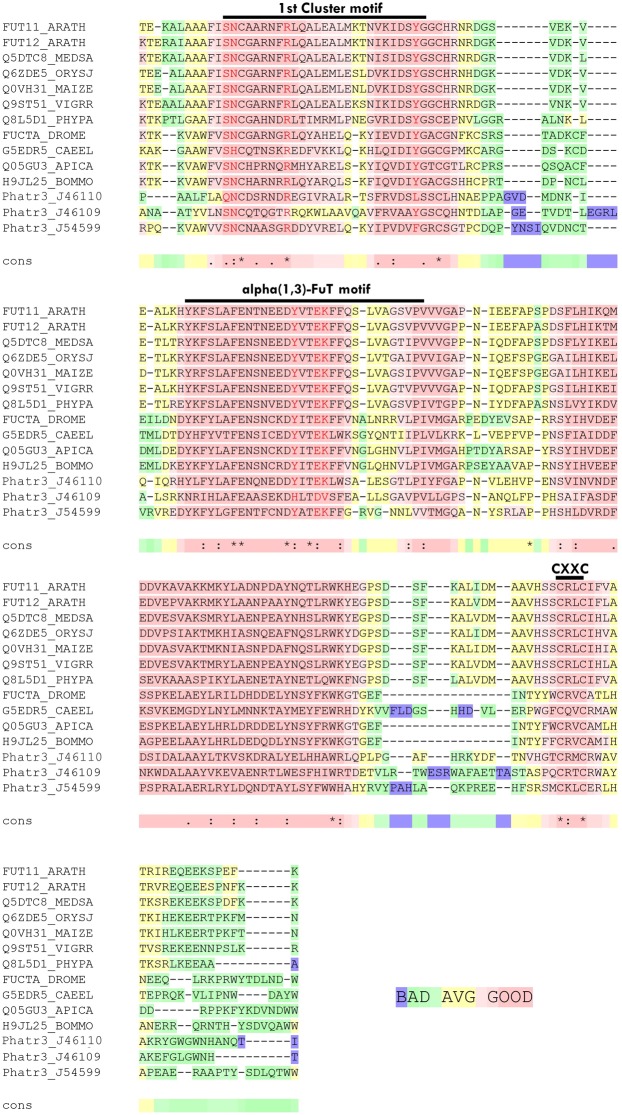
FIGURE 5.
Phaeodactylum tricornutum putative FuT exhibits strong amino acid identities with α(1,3)-fucosyltransferases. Amino acids sequences comparison with the T-coffee program (http://tcoffee.crg.cat/, Di Tommaso et al., 2011) of the C-terminal GT10 domains of the three putative fucosyltransferases encoded, respectively, by the Phatr3_J46109, Phatr3_J46110 and Phatr3_J54599 genes from Phaeodactylum tricornutum with biochemically characterized α(1,3)-fucosyltransferases from Arabidopsis thaliana (FUT11_ARATH and FUT12_ARATH), Medicago sativa (Q5DTC8_MEDSA), Oryza sativa (Q6ZDE5_ORYSJ), Zea mays Q0VH31_MAIZE), Vigna radiata (Q9ST51_VIGRR), Physcomitrella patens (Q8L5D1_PHYPA), Drosophila melanogaster (FUCTA_DROME), Caenorhabditis elegans (G5EDR5_CAEEL), Apis mellifera (Q05GU3_APICA) and Bombyx mori (H9JL25_BOMMO). The graphic output reflects the level of consistency of the alignment of a considered residue (from blue/green: badly or poorly supported to pink which corresponds to strongly supported). Conserved motifs of the GT10 domain are indicated on the top of the alignment.