Abstract
The Anti-Wolbachia (A·WOL) consortium at the Liverpool School of Tropical Medicine (LSTM) has partnered with the Global High-Throughput Screening (HTS) Centre at AstraZeneca to create the first anthelmintic HTS for neglected tropical diseases (NTDs). The A·WOL consortium aims to identify novel macrofilaricidal drugs targeting the essential bacterial symbiont (Wolbachia) of the filarial nematodes causing onchocerciasis and lymphatic filariasis. Working in collaboration, we have validated a robust high-throughput assay capable of identifying compounds that selectively kill Wolbachia over the host insect cell. We describe the development and validation process of this complex, phenotypic high-throughput assay and provide an overview of the primary outputs from screening the AstraZeneca library of 1.3 million compounds.
Keywords: high-throughput screening, phenotypic screening, neglected tropical diseases, Wolbachia, acumen, onchocerciasis, lymphatic filariasis
Introduction
Neglected tropical diseases (NTDs) are a major economic and health burden in developing countries across the world, affecting more than 1 billion people worldwide.1–3 This broad group of diseases encompasses a range of viral, bacterial, and parasitic infections, determined as neglected based on the lack of investment they receive in relation to the “big three” infectious diseases (tuberculosis, HIV/AIDS, and malaria).2 Despite the huge worldwide burden, this lack of investment has resulted in limited development of new drugs to treat these diseases.1,4 Although the veterinary drug moxidectin has recently been approved for the treatment of onchocerciasis, the lack of any new chemical entity approved for use against helminth infections in the past decade clearly highlights this issue.4,5
Lymphatic filariasis (LF) and onchocerciasis are two NTDs that are both caused by helminthic infections with filarial nematodes. These diseases are leading causes of global morbidity,2,6 but currently there are only three drugs suitable for mass drug administration (MDA): ivermectin (IVM), albendazole (ALB), and diethylcarbamazine citrate (DEC). The main limitation of these drugs is that they principally target the larval stage of these parasites (microfilariae).3,7 Microfilaricidal drugs interrupt transmission to the insect vectors and alleviate the symptoms of onchocerciasis. However, the adult parasites are able to repopulate microfilarial loads, requiring sustained long-term treatment delivery to cover their reproductive life span.8 This limitation is compounded by concerns over potential drug resistance9 and the risk of severe adverse events (SAEs) in settings co-endemic with Loa loa (IVM and DEC) and onchocerciasis (DEC).
An alternative approach, which overcomes these limitations, is to target the essential endosymbiont, Wolbachia, which has evolved a mutualism with the parasites that cause onchocerciasis and LF.10
A 4- to 6-week course of doxycycline eliminates the Wolbachia endosymbionts, leading to an effective macrofilaricidal (death of the adult worm) activity.11–14 Additionally, embryogenesis is blocked and microfilariae development is impaired in the vector,15 resulting in a complete block in transmission.3 Furthermore, as this bacterial target is not present in Loa loa, there is no risk of SAE, allowing for safe use in these co-endemic areas. Barriers to the wide-scale use of doxycycline as a macrofilaricidal treatment are the limitations of the long treatment period (4–6 weeks) and contraindications in pregnant women and children.
In the context of aiming to achieve the WHO targets of eliminating LF and onchocerciasis, Wolbachia as the drug target provides a promising new angle; however, new drugs are required (with no contraindications and a shorter treatment regime).2,3 The Anti-Wolbachia (A·WOL) consortium was established to meet this challenge. Recent drug discovery successes have relied heavily on collaboration and access to large compound libraries,1,4 with large pharmaceutical companies ever more open to partner in areas of clear unmet medical need.16–18
The complex biology and life cycle involved in LF and onchocerciasis has made industrial-scale drug discovery programs extremely challenging;19 however, the A·WOL consortium started to address these challenges through the development and deployment of an in vitro assay system employing an insect cell line stably infected with Wolbachia.2,7,20,21 Use of this system was central to the development of a drug discovery program that has already tested a variety of compound libraries from a range of sources, through multiple collaborations.7,21,22 This paper describes an innovative new collaboration between A·WOL and AstraZeneca that has allowed the first truly open-access collaboration between academia and industry, in the anthelmintic field.2,20 This partnership has taken collaborations a step further than the simple sharing of a compound library with a group already running screens against a particular molecular target or phenotype. Through this open-access collaboration scientists from A·WOL with knowledge of the disease biology and current screening assays against Wolbachia were given full access to the AstraZeneca compound library and laboratory facilities. This included access to specialist automation and internal expertise in the field of high-throughput screening (HTS). The collaboration culminated in the development, validation, and execution of a novel industrial-scale HTS against 1.3 million compounds, the largest of its kind ever completed against any anthelmintic NTD. Details of the completion, follow-up studies, and subsequent identification of five series of fast-acting macrofilaricides have been presented in Clare et al. (2019).23
Materials and Methods
Cell Culture
The C6/36 (wAlbB) cell line used in this screen has been described previously.2,7,20–23 In brief this mosquito (Aedes albopictus)-derived cell line is stably infected with Wolbachia pipientis (wAlbB). For use in the screen, cells were cultured in Leibovitz medium (Life Technologies, Loughborough, UK) supplemented with 20% fetal bovine serum (FBS; Fisher Scientific, Loughborough, UK), 2% tryptose phosphate broth (Sigma-Aldrich, Poole, UK), 1% nonessential amino acids (Sigma-Aldrich), and 1% penicillin-streptomycin (Sigma-Aldrich) at 26 °C, without additional CO2. The medium described was optimized through previous work.20
Large-Scale Cryopreserved Cell Bank
C6/36 (wAlbB) cells were cultured at scale in 16 × T225 cm2 flasks (Thermo Fisher, Waltham, MA) to generate 6.16 × 109 cells after 7 days of incubation. To produce the cryopreserved cell bank, spent medium was removed and replaced with 20 mL of fresh cell culture medium. The cells were detached by scraping, combined, and centrifuged. The pellet was resuspended in cryopreservation medium (90% FBS and 10% DMSO) to a density of 3 × 107 cells/mL. The cells were aliquoted 1 mL per cryovial for cryopreservation using a control rate freezer (Planar Kryo 560-16). This resulted in a cryopreserved cell bank of 190 vials.
Cell Bank Recovery and Quality Control for Screening
A single cryovial was recovered by defrosting at 37 °C for 45 s, followed by immediate resuspension in 40 mL of cell culture medium, resulting in a DMSO concentration of <0.25%. The cells were centrifuged and resuspended in 45 mL of cell culture medium in a T225 cm2 flask (VWR, Lutterworth, UK). After a 7-day incubation at 26 °C, with ambient CO2, the cells were tested (quality control [QC]) for Wolbachia infection levels by adding 40 μL of the resuspended cells to a 384-well, black, clear-bottom, tissue culture-treated plate (781090, Greiner Bio-One, Stonehouse, UK). Once settled, the cells were fixed for 20 min with formaldehyde (0.82% final concentration) supplemented with Hoechst 33342 (54 µg/mL final concentration, Life Technologies). A phosphate-buffered saline (PBS) wash was followed by incubation with SYTO 11 for 15 min (7.5 μM final concentration, Life Technologies). After a final PBS wash, the cells were left in PBS and analyzed on the PerkinElmer (Seer Green, UK) Operetta cellular imaging system, to assess the percentage of cells infected with Wolbachia. Confocal images were acquired using a 60× objective and the cell nuclei identified using the Hoechst stain, allowing for the cytoplasm (omitting the nucleus) to be located by the SYTO 11 staining. As previously reported by Clare et al. (2014),20 the cytoplasm texture was analyzed (with a more granular texture indicating a higher Wolbachia infection level). Based on image analysis of infected and uninfected cells, a threshold was set at a texture score of 0.0028, within the PerkinElmer Harmony analysis software. Above this threshold cells were classed as infected with Wolbachia, and those cultures with greater than 50% of the cell population infected were deemed to have passed QC and were subsequently used for screening.
Compound Handling
Compounds sourced from the AstraZeneca (Macclesfield, UK) library were plated from liquid stock (held at 10 mM in 100% DMSO and stored in 1536-well microtiter plates). For single-shot screening, 80 nL of this stock was dispensed into each well of a 384-well, clear-bottom microtiter plate (781090, Greiner Bio-One) via acoustic drop ejection (using Labcyte Echo 555 liquid handling units) to create assay-ready plates (ARPs). Once diluted through addition of 80 µL of cell suspension this would yield a final screening concentration of 10 µM. Onboard maximum/minimum controls were included on each ARP in two central columns, resulting in 16 wells of each control (containing 80 nL of either 100% DMSO or 5 mM doxycycline, respectively). Concentration–response curves for assay concordance testing and validation were also built using the Labcyte Echo 555 units, with a final dose range—following the addition of 80 µL of cell suspension—of 30 µM to 20 pM (across 13 points, using half-log dilutions). These compounds were selected and sourced from A·WOL compound libraries at a stock concentration of 10 mM in 100% DMSO. In order to create the 30 µM concentration point from a 10 mM stock, the volume of compound dispensed for these specific wells was increased to 240 nL (meaning that the DMSO concentration in the top dose points was three times higher than across the remainder of the compound curves).
Additional QC plates—containing only control wells—were interspersed into the screening runs. These ARPs included columns of “Max signal” (100% DMSO), “Min signal” (5 mM doxycycline), and “Reference signal” (50 µM doxycycline) repeated across the plate and were built in the same way as the ARPs, using the Labcyte Echo 555 units (80 nl of the concentration as described for each control).
Wolbachia Specific Primary Antibody Production
The antibody was made by Covance (Denver, CO) as previously described24,25 by immunizing five specific pathogen-free (SPF) rabbits with Wolbachia peptidoglycan-associated lipoprotein from Brugia malayi (wBmPAL) produced by New England BioLabs (NEB, Ipswich, MA). The antiserum was obtained and the anti-wBmPAL antibody purified by affinity column chromatography. From this we gained 350 mL of antibody, which we titrated against various concentrations of the secondary antibody, Alexa Fluor 680 goat anti-rabbit IgG (A-21076, far red fluorescence, Life Technologies).
Detection (acumen Explorer eX3 Analysis) and EnVision Read
The TTP acumen Explorer eX3 adherent cell cytometer (referred to from this point as “acumen”) was configured using a 633 nm excitation laser (via a 633 nm dichroic mirror) and Wolbachia fluorescence was detected by a 655 nm long pass filter (655–800 nm filter) (Suppl. Fig. S1). The acumen analysis software identified objects, based on separate areas of fluorescence, which were subsequently gated for excessive area and fluorescence intensity. The total area of the filtered objects (assumed to be cytoplasm-encapsulated Wolbachia) per well was calculated as the readout. This gave quantification of Wolbachia levels per well, where compounds with anti-Wolbachia activity were represented by a decreased total area per well.
As this decrease in area of Wolbachia fluorescence would also occur if the compounds showed host cell toxicity (low number of cells either with or without a Wolbachia infection), a cell toxicity read was included to identify such “false positives.” To optimize throughput, this toxicity read was carried out by a single fluorescent readout per well, based on the intensity of Hoechst staining, detected via a PerkinElmer EnVision plate reader. The EnVision was optimized using a bottom mirror with 355/103 nm excitation and 460/207 nm emission filters (set to 10 flashes per read). The acumen acquisition and analysis step was processed in approximately 20 min, while the EnVision read was obtained in 2 min. This total screening time of 22 min per plate allowed for a full run of 150 plates to be read in 55 h. To increase the throughput further, two acumen units and a single EnVision unit were utilized, allowing for the 150-plate batch to be read in 28 h.
Validation of the HTS Assay
The assay was robustly validated, following AstraZeneca’s established process.
AstraZeneca holds a validation set of 7000 compounds (representing the full library), which are run on at least two occasions against the assay (nominally termed occasion 1 and occasion 2). In addition, each occasion comprises two different physical picks of compounds from the AstraZeneca compound store. Each of these physical picks of compounds is plated in a different order and thus allows validation against inter- and intraplate variation within a given assay.
Each occasion of the AstraZeneca validation set testing took place on a separate experimental run at a single concentration (10 µM final concentration). Each experiment used a different cryopreserved cell population and freshly made reagents (mimicking a true experimental run as closely as possible). Alongside this “generic” validation set, 20 compounds were selected from the A·WOL drug discovery program, spanning various levels of both anti-Wolbachia activity and host cell toxicity. These compounds (including the well-characterized tetracycline, doxycycline) were run in concentration response. The IC50 values generated were compared to those calculated previously in imaging assays from A·WOL using a PerkinElmer Operetta.20–22
Screening Protocol
An overview of the 3-week screening protocol is presented in Supplemental Figure S2. On the morning of each screening run, cells recovered from a cryovial 7 days previously were QC checked for confluence and Wolbachia infection levels (as described earlier). The cells were counted on a Vi-Cell XR (Beckman Coulter, High Wycombe, UK) and diluted to a concentration of 25,000 cells/mL in assay medium (as culture medium described earlier). Compound plates were organized into batches of around 150, including 2 “max” plates (maximum signal, no compounds or DMSO added) at the beginning of each run and 3 QC plates (as described earlier) dispersed throughout the run. Using a Multidrop Combi (Thermo Scientific), 80 μL of cell suspension was added to each test well, resulting in a seeding density of 2000 cells per well. All plates were foil sealed using a PlateLoc thermal plate sealer (Velocity11, Menlo Park, CA) for 2 s at 230 °C, and incubated for 7 days at 26 °C, with ambient CO2. In addition to the screening plates, a single max plate was created with a clear seal in order to periodically monitor the cell growth over the 7 days, without having to disturb any assay plates.
On day 7 post cell dispense, the assay plates were loaded into the incubator (Liconic) on a BioCel 1800 automation system (Agilent Technologies, Santa Clara, CA) held at 26 °C, with ambient CO2. To enable the assay staining protocol, the BioCel system was configured with five Multidrop Combis (Thermo Scientific), two plate washers (PW 384, Tecan, Männedorf, Switzerland), three robotic arms, one plate carousel, one PlateLoc thermal plate sealer (Velocity11), and one automated plate seal remover (XPeel, Brooks Life Sciences, Irlam, UK) (Suppl. Fig. S3). Using this platform under the control of Agilent Vworks software, plates were allowed to enter the system every 2 min. The plates were fixed by adding 20 μL of 9% formaldehyde containing 3 µg/mL of Hoechst 33342 (Life Technologies) directly into each well containing 80 μL of cell culture, using one of the Multidrop Combis (1.8% final formaldehyde concentration and 0.6 µg/mL Hoechst), for 20 min. The fixative was aspirated off using the Tecan PW, and following this 80 μL of PBS was dispensed into each well using a Multidrop Combi. The PBS was aspirated off using the Tecan PW and the adherent cells were permeabilized by the addition of 80 μL of PBS + 0.25% Triton-X 100 (Sigma-Aldrich) using a Multidrop Combi. Following a 30 min incubation at room temperature with the permeabilization buffer, a further 80 μL of PBS wash was performed. The plates were blocked with the addition of 80 μL of blocking buffer (PBS + 6% bovine serum albumin [BSA] [Sigma-Aldrich]) for 40 min at room temperature, before removal of the blocking buffer by aspiration on the Tecan PW and addition of the anti-peptidoglycan-associated lipoprotein from the Wolbachia of the filarial nematode Brugia malayi (wBmPAL) primary antibody (in PBS + 6% BSA), using one of the Multidrop Combis (30 µL of 1:2000 dilution in blocking buffer). Primary antibody was incubated for 12 h at room temperature, before unbound antibody was removed by aspiration on the Tecan PW followed by addition of 80 µL of wash buffer (PBS + 0.05% polysorbate [Sigma-Aldrich]) and incubation at room temperature for 6 min. Following aspiration of the wash buffer, addition of the Alexa Fluor 680 goat anti-rabbit (A-21076, Life Technologies) secondary antibody (30 µL of 1:400 dilution in blocking buffer) was performed with a third Multidrop Combi and plates were incubated for 1 h at room temperature. Following this incubation, the secondary antibody was removed with 2× 80 µL, 6 min PBS + 0.05% polysorbate washes (as described earlier for the primary antibody) and plates left with 40 μL of PBS per well. As a final step, the assay plates were foil sealed (using the PlateLoc at 170 °C for 1 s) and output to stacker units on the BioCel platform (Suppl. Fig. S3).
The fixed antibody-stained plates were read on a PerkinElmer EnVision (Hoechst host cell toxicity analysis) and subsequently a TTP acumen (Wolbachia analysis), both automated via a HighRes Biosolutions Dual pod six-sided MicroStar under the control of Cellario scheduling software (Suppl. Fig. S4).
Data Analysis
Genedata Screener was used for all data analysis. Readouts from both the acumen and EnVision were imported into Screener, in which data for toxicity (Hoechst read on the EnVision) and anti-Wolbachia activity (Wolbachia area detected on the acumen) were normalized on an individual plate basis. For single-concentration primary screening, the toxicity read was normalized to the median read from all compounds on each plate as it is assumed that (1) toxic compounds are randomly distributed over the plates in a given screening run, and (2) toxicity against the insect cell line should be limited within the AstraZeneca collection. The plate median is therefore taken to represent a “Max Hoechst signal” (0% toxicity toward the host insect cell). The anti-Wolbachia reads were normalized against the Max signal (DMSO control wells) and Min signal (5 μM doxycycline wells)—taken to represent 0% and 100% anti-Wolbachia activity, respectively.
The data were processed to identify “hits” (anti-Wolbachia activity) that presented little or no toxicity to the host cell line. To do this, the normalized anti-Wolbachia data were cut at ≥80% inhibition and the compounds falling above this cut were further annotated to identify those with ≤60% cytotoxic effect on the C6/36 host cell line.
Additionally, all plates were monitored for Z prime robust Z score, and signal to background. These data were analyzed on a per-plate basis within each run so we could flag up and repeat plates that did not pass our acceptance criteria, but also across all runs in the completed screen to follow reliability and identify any potential adverse data trends.
Results and Discussion
To establish a screening assay suitable for the delivery of a HTS campaign, we built upon the shared knowledge and experience of both A·WOL and AstraZeneca. Taking the C6/36 (wAlbB) cell line and Wolbachia specific antibody described previously,24,25 we built and validated an assay on the TTP acumen Explorer eX3 adherent cytometer platform. This platform has many advantages over traditional imaging platforms in the high-throughput setting (primarily driven by speed of whole-well acquisition, combined with low data burden per scan). In the context of this screen, we did not require the resolution of a true imaging platform in order to give us spatial information, but rather needed a gross measure of Wolbachia number across the whole test well.
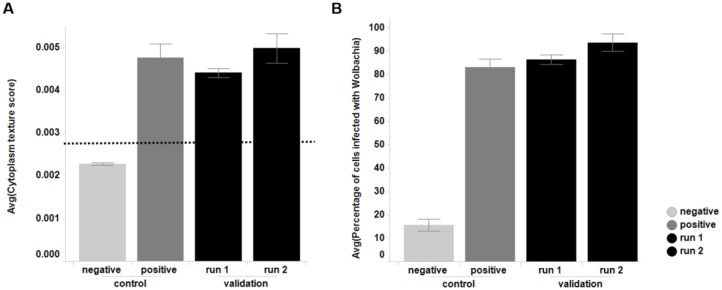
Although the C6/36 (wAlbB) line has been used for smaller-scale screening activities previously within A·WOL, experience with this cell line has also shown that Wolbachia infection levels can fluctuate over extended periods of continuous culture. For this reason, we invested time early in the assay development process, to produce and validate a large-scale cryopreserved cell bank (sufficient to complete screening of the full compound collection). The subsequent cell bank used for screening was consistent over the >30 experimental runs needed to complete the HTS campaign and therefore allowed for true comparison of compound activity from run to run. As described in the methods as part of the screening protocol, cells were recovered from cryovials and grown for a week, before an imaging-based QC step to determine Wolbachia infection levels for screening. Figure 1 shows the consistency (in terms of Wolbachia infection levels) from two independent cryovial recoveries and is representative of the consistency seen across the entire screen.
Figure 1.
Control samples (with positive and negative Wolbachia infection) and QC results for the cells used in the validation of this assay, sourced from the single large-scale cryopreserved cell batch used for the full screen. (A) Cytoplasm texture score. The dotted line represents the threshold of the texture score above which cells are classed as infected (0.0028). (B) The percentage of cells that are classed as infected based on their cytoplasm texture score being above 0.0028. Comparison of the negative control with all Wolbachia-infected samples (positive control, validation runs 1 and 2) gives a Z prime of 0.67 and a signal to background of 5.6.
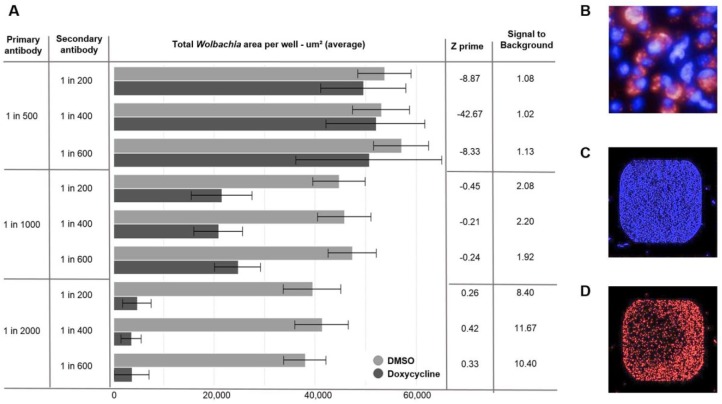
With the cellular system established, we were able to begin optimization of the detection endpoints to be used within the screen. As previously mentioned, key for this project was to find chemical equity that displayed anti-Wolbachia activity over a 7-day treatment window, but with negligible cytotoxic effect on the host C6/36 cells. In order to generate this multiparametric data at the primary screening stage, we established an assay using two markers: (1) Hoechst to stain DNA and enable quantification of C6/36 host cells (2) anti-wBmPAL (detected through an Alexa 680 secondary antibody) as a specific marker of Wolbachia. Using a specific marker of Wolbachia as opposed to the SYTO 11 protocol described previously20,21 enabled assay transfer to the acumen platform. While losing resolution from the confocal imaging method used previously at A·WOL, the acumen detection enabled a much faster acquisition time per plate (reducing from 3 h to 20 min), with the additional benefit of whole-well scanning, rather than selected fields of view. This improved acquisition time made the full collection screen achievable within an acceptable time frame. Both Alexa 488 (green fluorescence) and Alexa 680 (far red fluorescence) conjugated secondary antibodies were tested as part of this assay development; however, we opted to move forward with the red-shifted fluorochrome in detection of the Wolbachia, in order to ensure maximum separation from the Hoechst emission spectra (461 nm). Past experience of fluorescent detection modalities applied to diversity screening has shown that higher-wavelength fluorochromes are preferable as they are farther away from the blue/green part of the spectra, where many cyclic molecules fluoresce.26 Experience within AstraZeneca (unpublished) has also shown that the artifacts generated through compound crystalization within imaging assays can be minimized by moving to higher wavelengths. A matrix of various concentrations of both primary and secondary antibody was tested in order to reduce the antibody concentrations where possible, allowing for a balance between cost and high-quality signal. Based on this optimization, a 1:2000 dilution of the primary antibody combined with a 1:400 dilution of the secondary antibody was chosen for the screen ( Fig. 2 ), as this combination gave both the best Z prime and signal-to-background window.27
Figure 2.
(A) Wolbachia total area read on the acumen for matrix titration of primary and secondary antibody concentrations. The columns on the right include analysis of the titrations based on comparison of results from DMSO (Wolbachia positive control) and 5 µM doxycycline (Wolbachia negative control) treatment. (B) Operetta image (60× wide-field) of C6/36 (wAlbB) cells stained with Hoechst to identify the nuclei (blue) and antibody staining (primary antibody against wBmPAL with Alexa Fluor 680 secondary antibody) to identify the Wolbachia (red). (C) C6/36 (wAlbB) cells stained with Hoechst (blue) acquired on the acumen. (D) C6/36 (wAlbB) cells stained with Wolbachia specific antibody (red) imaged on the acumen.
As further optimization of the screening protocol, we addressed both the wash regime used in the antibody staining and incubation conditions for the wBmPAL primary antibody (minimizing timing for each step wherever possible).
Further miniaturization of this assay was limited by the long-running challenge of achieving robust and consistent plate washing in a 1536-well format. Due to the multiple wash steps required within this assay, although we did explore newer technologies amenable to 1536-well washing (namely, the BlueCat centrifugal washer), we opted to remain in 384-well format in order to preserve consistency and accuracy.
Even though it was possible to generate both the specific Wolbachia readout and the Hoechst readout on the acumen, these reads would need to run sequentially on each well (due to excitation with different lasers). This was considered an approach to screening; however, we quickly discounted this due to the time penalty incurred. Due to the consistency in cell number achieved through use of a single cryobank along with automated assay build (coupled to the intensity and consistency of Hoechst staining), we found that we were able to utilize a different modality for establishing a measured cell number, using the PerkinElmer Envision plate reader. Hence our finalized screening assay consisted of two independent readouts (on two independent platforms). This was made possible through the application of industrial-scale laboratory robotics (as described above and in the supplemental section).
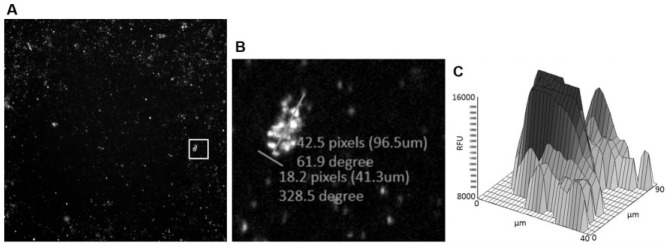
The early decision with regard to the detection system of choice meant that assay development could be tailored to the acumen unit from the start. Test plates were imaged on the PerkinElmer Operetta during assay development (to aide understanding through highly resolved images) along with scanning on the acumen. This combined approach allowed fast decision making on population gating to apply to the acumen on-the-fly analysis. The screening assay is not configured with high enough resolution to distinguish individual Wolbachia objects, and therefore objects needed to be gated based on maximum cluster size. Through the use of imaging during assay development, we determined that the largest Wolbachia clusters were around 100 μm × 40 μm; therefore, an upper gate was set on the acumen for object area (detected using the 680 nm channel) at 4000 μm2 ( Fig. 3 ).
Figure 3.
(A) Operetta image (4× objective) of 384-well plate containing C6/36 (wAlbB) cells infected with Wolbachia. Primary antibody against wBmPAL with Alexa Fluor 680 secondary antibody. White box denotes Wolbachia clump. (B) Zoomed view into area denoted by white box in (A) showing Wolbachia clump with measuring lines. (C) Corresponding acumen 2D fluorescence intensity plot of a Wolbachia clump.
Once antibody staining and wash steps had been optimized for detection on the acumen platform, the resultant protocol was transferred onto an Agilent BioCel 1800 platform and configured as described earlier (Suppl. Fig. S3), enabling fully automated assay construction. Several rounds of optimization were undertaken in order to establish the optimum timing for plate entry onto the platform (otherwise known as “pacing time” or “tick time” for the protocol). The complexity of the overall assay procedure resulted in a finalized three-stage assay (Suppl. Fig. S2):
Manual 1-week recovery of cryopreserved cells in T225 cm2 flasks, followed by addition on day 7 into assay-ready test plates using a Multidrop Combi (Thermo Scientific) inside a class II biosafety cabinet, to ensure product protection.
On day 14, after 7 days of incubation with compounds, formaldehyde fixation, Hoechst staining, and primary antibody staining of test plates was conducted on the Agilent BioCel 1800 unit (local exhaust ventilation extracted for safety reasons, to prevent a localized buildup of formaldehyde vapor). On day 15, following a 12 h incubation of the primary antibody on the first plate, the second part of the BioCel 1800 protocol was initiated to carry out secondary antibody staining of the test plates.
From days 15 to 17, depending on the number of test plates in the batch, the plates were transferred to the HighRes Microstar platform, where they were sequentially read on a PerkinElmer Envision (to capture the Hoechst endpoint), followed by acquisition of the Wolbachia area endpoint on the acumen. The acumen acquisition was split over two individual readers pooled on the HighRes Microstar platform in order to maintain throughput. This was followed by analysis using Genedata Screener as the data became available.
Due to the complexity of the automated protocol and the fact that assay runs were interleaved (in order to progress through the compound collection in a timely fashion), it was important to consider error recovery strategies. Assay plates had been shown to remain relatively stable if left for extended periods in PBS during the protocol; however, deterioration in assay quality was observed if fixative or permeabilization steps extended beyond the optimized time. The automated part of the protocol was configured so that a limited number of assay plates were released onto the robotic system at any one time (effectively forming a series of mini-batches). This phased-release strategy was employed in order to mitigate against potential large-scale loss of test plates due to unforeseen automation crashes.
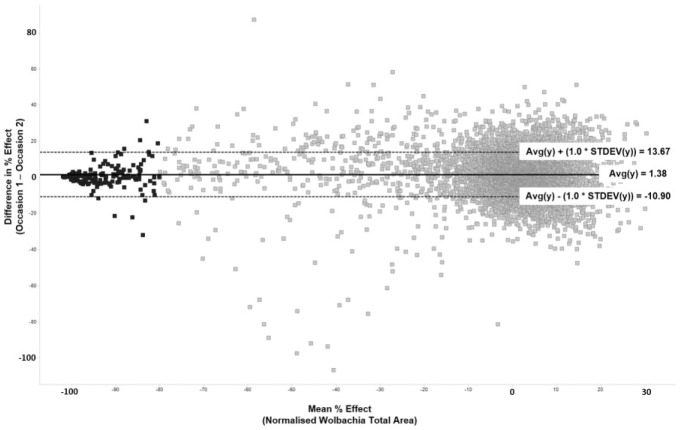
With the assay system established, along with the screening procedure (including all automation protocols), validation of the assay for HTS was performed as described earlier. Validation showed the assay to perform well and established good agreement between occasion 1 and occasion 2 of the validation compound set. The mean of the differences in anti-Wolbachia activity fell close to the zero line when plotted as a Bland-Altman28 (difference) plot with ±1 standard deviation lines at less than 14% variance ( Fig. 4 ).
Figure 4.
Bland-Altman plot representing the variance in anti-Wolbachia activity between occasion 1 and occasion 2 testing the AstraZeneca validation set of compounds. The average difference in percent effect between paired results lies close to 0 (1.38) and 1*standard deviation is less than 14%. Darker colored points represent those with average activity ≥80% (the cutoff for active compounds in the screen).
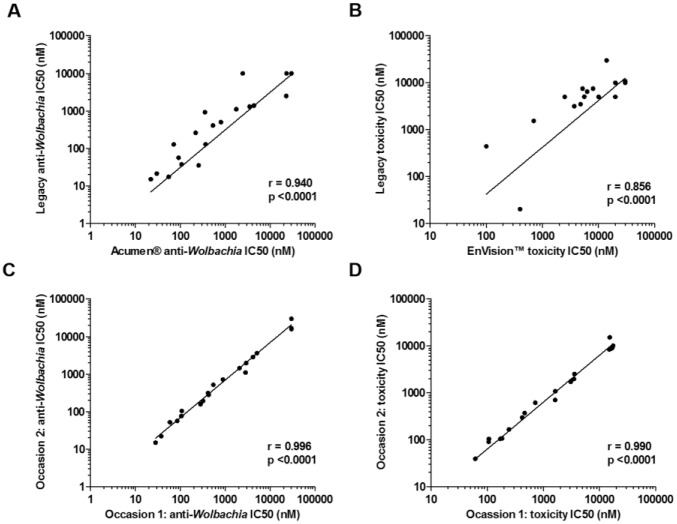
In addition to the validation data against the AstraZeneca validation set, concordance with IC50 data generated historically from A·WOL compounds was very good ( Fig. 5A–D ), building further confidence in the assay as robust enough to move to a full HTS campaign. Figure 5A shows a strong agreement in anti-Wolbachia IC50 values between the acumen assay and legacy A·WOL assays (Spearman two-tailed correlation—where a coefficient [r value] of 1 is a perfect agreement; r = 0.9405, p < 0.0001). Doxycycline reported consistent IC50 values of 17.8 nM in the acumen assay (mean, n = 14 spread across two separate experimental runs) and 16 nM from A·WOL legacy assays (median from 396 assays). Figure 5B shows a similar strong agreement in host cell toxicity between the A·WOL legacy data and those from the EnVision (r = 0.856, p < 0.0001). To add further confidence to the robustness of the screening assay, Figure 5C,D shows the statistical agreement of the IC50 values over 2 different experimental occasions (r > 0.99, p < 0.0001).
Figure 5.
Concordance testing of IC50 values for anti-Wolbachia activity and host cell toxicity across a set of 20 compounds from the A∙WOL drug discovery program. (A) Anti-Wolbachia IC50 values in A·WOL legacy screens versus acumen screen at AstraZeneca. (B) Toxicity IC50 values in legacy versus EnVision screen at AstraZeneca. (C) acumen anti-Wolbachia IC50 values for occasion 1 versus occasion 2 in the AstraZeneca screen. (D) EnVision toxicity IC50 values for occasion 1 versus occasion 2 in the AstraZeneca screen.
With each plate taking 22 min to read on the acumen (Wolbachia analysis) following the read on the EnVision (toxicity analysis), the planned batch size of 100–150 plates per day would take approximately 28 h to complete—with two acumen units employed in a pooled fashion on the HighRes Microstar automation platform. In order to carry out multiple runs per week (ideally four) and to allow for downtime, it was important to establish stability of the assay signal over an extended time frame. The AstraZeneca validation set of compounds (described earlier) was rescanned, 7 days after the initial read (having stored the plates sealed at 4 °C). The Wolbachia signal remained consistent over this 7-day period (Suppl. Figs. S5 and S6).
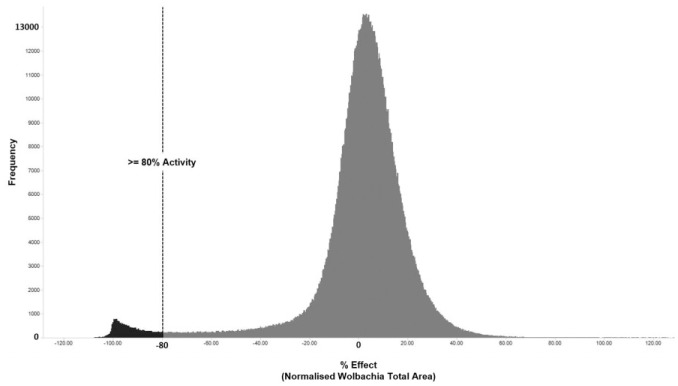
The development of a HTS-compatible phenotypic assay enabled the screening of ~1.3 million compounds from the AstraZeneca collection with a resultant hit rate of 2.36% ( Fig. 6 )—following annotation against the Hoechst measure of ≤60% cytotoxic effect on the C6/36 host cell line the final hit rate was reduced to 1.56% (20,255 compounds). These hits were clustered by structure and rationalized down before subsequent confirmation, though the assay described here is in IC50 format and progressed through further cascade assays (both within AstraZeneca and A·WOL) in order to build more annotation against the chemistry. Details of the completion, follow-up studies, and subsequent identification of five series of fast-acting macrofilaricides have been presented in Clare et al. (2019).23
Figure 6.
Frequency plot of primary HTS results across the 1.3 million compounds screened. The dotted line shows the cutoff used in the screen to select hits (≥80% activity as measured by total Wolbachia area). The distribution can be seen centered around zero with a tail leading out toward the left (active compounds). The proportion of compounds falling beyond the cutoff line shown is 2.36% of the total compounds tested.
The combination of academic expert knowledge in a particular disease area with expertise in industrial-scale HTS is a model that other groups are now beginning to implement; however, the tight integration of A·WOL staff embedded within the HTS Department at AstraZeneca for the duration of the screen optimization, validation, and ultimate execution is, to our knowledge, a step further than other partnerships. This collaborative public–private venture is certainly unique in the NTD space, and we believe it has brought unique learning to both sides.
The Discovery Sciences Department at AstraZeneca has a leading position as a group delivering the skills and capabilities essential for early-phase drug discovery on an industrial scale, integrated within one global department. The A·WOL consortium has a leading position in the field of research against anti-Wolbachia agents, with specific biological tools such as the C6/36 (wAlbB) Wolbachia-infected cell line being instrumental to the HTS described. Combining the skills, resources, and expertise of both organizations was required in order to build, validate, and execute the complex screen described here.
The output from the screening activity described has identified novel chemical starting points from the AstraZeneca compound collection, resulting in the identification of five series of novel fast-acting macrofilaricides,23 allowing for the potential of an improved medication against both LF and onchocerciasis. These starting points for discovery will form the basis of future publications specifically addressing the discovery process from hit through to leads and optimized drug candidates.21,22
Supplemental Material
Supplemental material, 3._ClareClark_Supplemental for Development of a High-Throughput Cytometric Screen to Identify Anti-Wolbachia Compounds: The Power of Public–Private Partnership by Rachel H. Clare, Roger Clark, Catherine Bardelle, Paul Harper, Matthew Collier, Kelly L. Johnston, Helen Plant, Darren Plant, Eileen McCall, Barton E. Slatko, Lindsey Cantin, Bo Wu, Louise Ford, David Murray, Kirsty Rich, Mark Wigglesworth, Mark J. Taylor and Stephen A. Ward in SLAS Discovery
Acknowledgments
The authors would like to thank the members of the Global HTS Department at AstraZeneca for its continued support over the course of the screen execution, and members of the Sample Management Group at AstraZeneca for provision of the assay-ready screening plates. We would also like to thank Dr. W. David Hong for the selection of the set of validation compounds from the A.WOL drug discovery program.
Footnotes
Supplemental material is available online with this article.
Declaration of Conflicting Interests: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: C.B., P.H., M.C., H.P., D.P., E.M., D.M., and M.W. are employees and therefore shareholders of AstraZeneca; however, they have no financial or intellectual property rights to the outputs presented in this manuscript, which were transferred to the Liverpool School of Tropical Medicine (LSTM), a charitable organization. The remaining authors declare no competing interests.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The project was funded by AstraZeneca and A·WOL, with the latter supported by a grant from the Bill & Melinda Gates Foundation to the LSTM. Antibody production was funded by NEB.
ORCID iD: Rachel H. Clare  https://orcid.org/0000-0002-3945-0530
https://orcid.org/0000-0002-3945-0530
References
- 1. Burrows J. N., Elliott R. L., Kaneko T., et al. The Role of Modern Drug Discovery in the Fight against Neglected and Tropical Diseases. MedChemComm 2014, 5, 688–700. [Google Scholar]
- 2. Johnston K. L., Ford L., Taylor M. J. M. Overcoming the Challenges of Drug Discovery for Neglected Tropical Diseases: The A·WOL Experience. J. Biomol. Screen. 2014, 19, 335–343. [DOI] [PubMed] [Google Scholar]
- 3. Slatko B. E., Luck A. N., Dobson S. L., et al. Wolbachia Endosymbionts and Human Disease Control. Mol. Biochem. Parasitol. 2014, 195, 88–95. [DOI] [PubMed] [Google Scholar]
- 4. Panic G., Duthaler U., Speich B., et al. Repurposing Drugs for the Treatment and Control of Helminth Infections. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 185–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Opoku N. O., Bakajika D. K., Kanza E. M., et al. Single Dose Moxidectin versus Ivermectin for Onchocerca volvulus Infection in Ghana, Liberia, and the Democratic Republic of the Congo: A Randomised, Controlled, Double-Blind Phase 3 Trial. Lancet 2018, 392, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taylor M. J., Hoerauf A., Townson S., et al. Anti-Wolbachia Drug Discovery and Development: Safe Macrofilaricides for Onchocerciasis and Lymphatic Filariasis. Parasitology 2014, 141, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnston K. L., Ford L., Umareddy I., et al. Repurposing of Approved Drugs from the Human Pharmacopoeia to Target Wolbachia Endosymbionts of Onchocerciasis and Lymphatic Filariasis. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Turner H. C., Walker M., Churcher T. S., et al. Modelling the Impact of Ivermectin on River Blindness and Its Burden on Morbidity and Mortality in African Savannah: EpiOncho Projections. Parasites Vectors 2014, 7, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Slatko B. E., Taylor M. J., Foster J. M. The Wolbachia Endosymbiont as an Anti-Filarial Nematode Target. Symbiosis 2010, 51, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taylor M. J., Voronin D., Johnston K. L., et al. Wolbachia Filarial Interactions. Cell. Microbiol. 2013, 15, 520–526. [DOI] [PubMed] [Google Scholar]
- 11. Hoerauf A., Specht S., Büttner M., et al. Wolbachia Endobacteria Depletion by doxycycline as Antifilarial Therapy Has Macrofilaricidal Activity in Onchocerciasis: A Randomized Placebo-Controlled Study. Med. Microbiol. Immunol. 2008, 197, 295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turner J. D., Tendongfor N., Esum M., et al. Macrofilaricidal Activity after Doxycycline Only Treatment of Onchocerca volvulus in an Area of Loa loa Co-Endemicity: A Randomized Controlled Trial. PLoS Negl. Trop. Dis. 2010, 4, e660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taylor M. J., Makunde W. H., McGarry H. F., et al. Macrofilaricidal Activity after Doxycycline Treatment of Wuchereria bancrofti: A Double-Blind, Randomised Placebo-Controlled Trial. Lancet 2005, 365, 2116–2121. [DOI] [PubMed] [Google Scholar]
- 14. Walker M., Specht S., Churcher T. S., et al. Therapeutic Efficacy and Macrofilaricidal Activity of Doxycycline for the Treatment of River Blindness. Clin. Infect. Dis. 2015, 60, 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Albers A., Esum M. E., Tendongfor N., et al. Retarded Onchocerca volvulus L1 to L3 Larval Development in the Simulium damnosum Vector after Anti-Wolbachial Treatment of the Human Host. Parasites Vectors 2012, 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clark R., Harper P., Wigglesworth M. Technological and Sociological Advances in HTS: Evolution and Revolution? Eur. Pharm. Rev. 2013, 18, 55–60. [Google Scholar]
- 17. Mullard A. European Lead Factory Opens for Business. Nat. Rev. Drug Discov. 2013, 12, 173–175. [DOI] [PubMed] [Google Scholar]
- 18. Lessl M., Schoepe S., Sommer A., et al. Grants4Targets—An Innovative Approach to Translate Ideas from Basic Research into Novel Drugs. Drug Discov. Today 2011, 16, 288–292. [DOI] [PubMed] [Google Scholar]
- 19. De Rycker M., Baragaña B., Duce S. L., et al. Challenges and Recent Progress in Drug Discovery for Tropical Diseases. Nature 2018, 559, 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clare R. H., Cook D. A., Johnston K. L., et al. Development and Validation of a High-Throughput Anti-Wolbachia Whole-Cell Screen: A Route to Macrofilaricidal Drugs against Onchocerciasis and Lymphatic Filariasis. J. Biomol. Screen. 2014, 20, 64–69. [DOI] [PubMed] [Google Scholar]
- 21. Johnston K. L., Cook D. A., Berry N. G., et al. Identification and Prioritization of Novel Anti-Wolbachia Chemotypes from Screening a 10,000-Compound Diversity Library. Sci. Adv. 2017, 36, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hong W. D., Benayoud F., Nixon G. L., et al. AWZ1066S, a Highly Specific Anti-Wolbachia Drug Candidate for a Short-Course Treatment of Filariasis. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 1414–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clare R. H., Bardelle C., Harper P., et al. Industrial Scale High-Throughput Screening Delivers Multiple Fast Acting Macrofilaricides. Nat. Commun. 2019, 10, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Turner J. D., Langley R. S., Johnston K. L., et al. Wolbachia Lipoprotein Stimulates Innate and Adaptive Immunity through Toll-Like Receptors 2 and 6 to Induce Disease Manifestations of Filariasis. J. Biol. Chem. 2009, 284, 22364–22378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnston K. L., Wu B., Guimarães A., et al. Lipoprotein Biosynthesis as a Target for Anti-Wolbachia Treatment of Filarial Nematodes. Parasites Vectors 2010, 3, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thorne N., Auld D. S., Inglese J. Apparent Activity in High-Throughput Screening: Origins of Compound-Dependent Assay Interference. Curr. Opin. Chem. Biol. 2010, 14, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J. H., Chung T. D. Y., Oldenburg K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [DOI] [PubMed] [Google Scholar]
- 28. Bland J. M., Altman D. G. Statistical Methods for Assessing Agreement between Two Methods of Clinical Measurement. Lancet 1986, 327, 307–310. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, 3._ClareClark_Supplemental for Development of a High-Throughput Cytometric Screen to Identify Anti-Wolbachia Compounds: The Power of Public–Private Partnership by Rachel H. Clare, Roger Clark, Catherine Bardelle, Paul Harper, Matthew Collier, Kelly L. Johnston, Helen Plant, Darren Plant, Eileen McCall, Barton E. Slatko, Lindsey Cantin, Bo Wu, Louise Ford, David Murray, Kirsty Rich, Mark Wigglesworth, Mark J. Taylor and Stephen A. Ward in SLAS Discovery