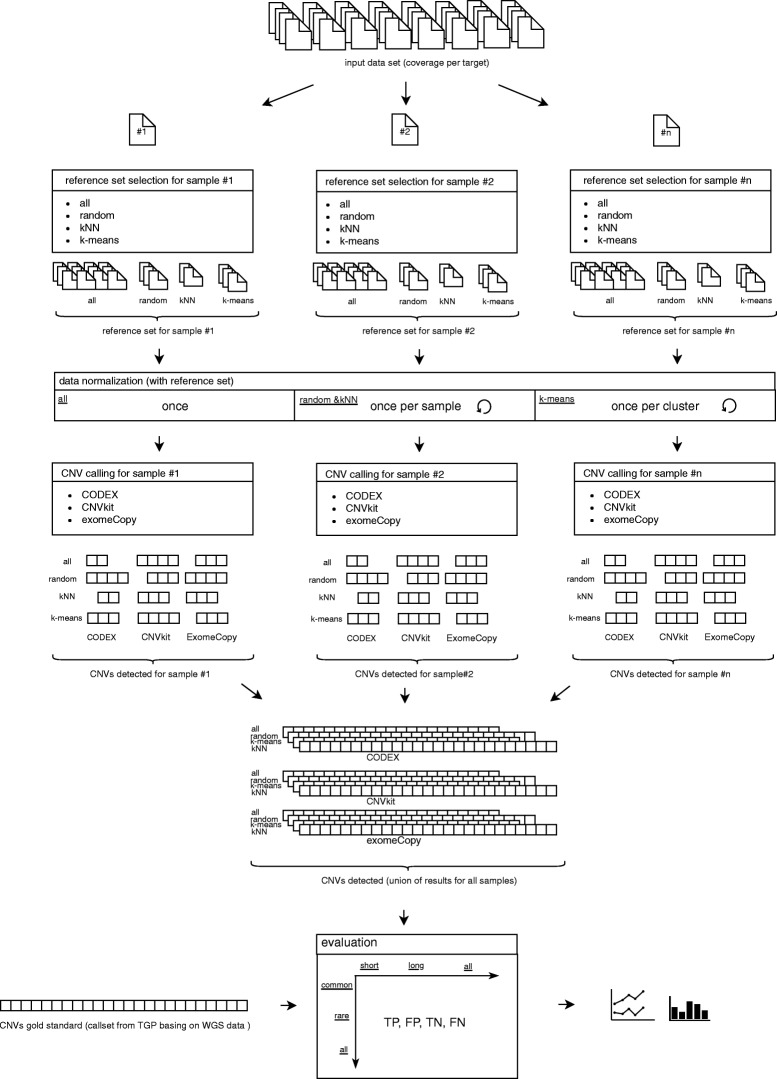
Fig. 2.
Workflow of the research method presented in the study. The input dataset is a set of numbers which depict the depth of coverage in samples on specified exons. Each sample from this dataset is processed by a reference sample set selector module, which is responsible for designating a set of samples that will be the reference collection. As a consequence, every element from the input dataset has its own, independent reference set. The normalization step uses the determined reference sets to perform normalization. This step is performed only once for ”all” method, once per sample in “kNN” and “random” approach and once per cluster in “k-means” strategy. Then, for each generated sample, we apply CNV calling performed by three callers: exomeCopy, CODEX and CNVkit. The input for the CNV detecting tool is a set of samples consisting of the investigated sample and its reference panel. After calling CNVs, the events in the investigated sample are filtered and, this set of events is added to the final set of CNVs. Having processed all samples from the input dataset, the union of all partial per-sample results stored as the output call set for each approach combining selection method and variant caller. The evaluation of the results is performed against CNVs call set gold standard, delivered by 1000 Genomes Project. Variations are additionally categorized into common and rare as well as short and long categories, which allows us to precisely calculate the True Positives, True Negatives, False Positives and False Negatives metrics in those groups