Abstract
Background
Gut bacteria are an important component of the microbiota ecosystem in humans and other animals, and they play important roles in human health. The aim of this study was to investigate the relationship between gut microbiota and multiple demographical-, behavioral-, or biochemical-related factors in subjects with chronic disease. Subjects with a very wide age range who participated in community-based chronic disease prevention and screening programs in China were enrolled. We analyzed the intestinal microbiota composition using 16S rRNA-based high-throughput sequencing of fecal samples, analyzed the association between gut microbiota structure and multiple demographical, behavioral, and biochemical factors, and compared the differences in microbiota composition in age-stratified groups with different blood glucose levels.
Results
Our results showed that both age and blood glucose levels had a significant impact on the gut microbiota structure. We also identified several taxa showed distinct abundance in groups with different glucose levels. Lactobacillus and Bifidobacterium at genus level and their related taxa were more abundant in the GLU high group comparing with GLU normal group and in NGR group comparing with DM group. Further analysis using the age-stratified data showed that blood glucose levels had a more significant impact on the gut microbiota in the ≥76 y age group than in the ≤75 y age group, which indicated that it is necessary to take age into account when conducting such studies. Moreover, we identified several taxa that were highly associated with blood glucose levels in the ≥76 y age group but not in the ≤75 y age group. Within the ≥76 y age group, Lachnospiraceae incertae sedis and Bacteroides were more abundant in the GLU normal group, whereas Lactobacillus and Bifidobacterium at genus level were more abundant in the GLU high group.
Conclusions
This result suggested that taxa that are capable of differentiating blood glucose levels might differ significantly in different age groups.
Keywords: Gut microbiota, Age, Blood glucose level, 16S rRNA-based high-throughput sequencing
Background
Diabetes is a common metabolic disease characterized by hyperglycemia resulting from defects in insulin secretion, insulin action, or both. [1]. Worldwide, 415 million people live with diabetes, and an estimated 193 million people have undiagnosed diabetes. China is on pace to become the country with the highest population of diabetics in the world, with 103 million people diagnosed. Type 2 diabetes (T2D) accounts for more than 90% of patients with diabetes [2] and leads to microvascular and macrovascular complications that cause blindness, kidney failure, lower limb amputation, etc. The care and treatment of diabetics places considerable socioeconomic pressures on the medical system.
Gut bacteria are an important component of the microbiota ecosystem in humans and other animals, and they play important roles in human health, such as nutrient absorption, homeostatic control of energy balance, immunoregulation, gastrointestinal development, and many other physiological processes. Gut bacteria can mirror host physiology [3]. Gastrointestinal microbiota represent a complex ecosystem with enormous diversity. Gut microbiota not only participate in the synthesis of dietary fatty acids and the absorption of fat-soluble vitamins, but also affect the colonization of pathogenic bacteria and regulate bile acid transformation, consequently regulating energy homeostasis [4–6]. The diversity and composition of gut microbiota are also affected by host physiological factors, hereditary factors, and dietary and environmental factors [7, 8]. Dysbiosis of gut bacterial communities is associated with many chronic diseases, such as type I diabetes (TID), obesity, inflammatory bowel disease (IBD), rheumatoid arthritis, cancer, autism, and allergies [9]. Several studies have provided evidence that the pathogeneses of type 2 diabetes (T2D), such as chronic low-grade inflammation and insulin resistance, are significantly associated with intestinal microbiota compositional changes, which cause increased absorption of monosaccharides, increased production of insulin resistance-related substances, changes in intestinal lining permeability, and increased production of lipopolysaccharides (LPS) [10–12]. The differences between the composition of the intestinal microbiota in humans with T2D and non-diabetic persons [10, 13–20] indicate that T2D is associated with differences in Actinomycetaceae, Alistipes, Bacteroides, Betaproteobacteria, Bifidobacterium, Clostridia, Coriobacteriaceae, Desulfovibrionaceae, Erysipelotrichaceae, Eubacterium, Faecalibacterium, Firmicutes, Fusobacterium, Lachnospiraceae, Lactobacillus, Parabacteroides, Peptostreptococcaceae, Planococcaceae, Prevotella, Propionibacteriaceae, Proteobacteria, Roseburia, Streptococcus, Veillonellaceae, and Verrucomicrobia. These studies provide evidence of an association between intestinal dysbiosis and T2D. However, there are great discrepancies across studies with respect to taxa changes in T2D patients as compared with healthy controls. This inconsistency could be explained by different sequencing technologies, different statistical methods, and the selection of thresholds of significance. However, ineffective control of confounding factors that might affect the association between microbiota and the target population also contributes to the observed inconsistency. It is evident that the activity and composition of the gut microbiota change with advancing age [21]. Aging is considered a chronic inflammation process [22], and dysbiosis plays a pivotal role in the pathogenesis and development of age-related diseases including T2D [23]. However, to our best knowledge, studies evaluating the effects of age-related factors on the relationship between intestinal microbiota and T2D are lacking.
In this study, we investigated the relationship between gut microbiota and plasma glucose levels, age, as well as multiple demographical-, behavioral-, and biochemical-related factors in subjects with a very wide age range who participated in community-based chronic disease prevention and screening programs in China. In addition, we compared the composition of gut microbiota in groups with different plasma glucose levels after age stratification.
Results
Characteristics of the participants
Samples from a total of 133 participants including 55 males and 78 females were investigated in the study. The major demographic, clinical, and behavioral characteristics of the participants are shown in Table 1. The age of the participants ranged from 44 to 88. According to WHO diagnostic criteria for diabetes (1999), 78 participants who had fasting plasma glucose < 6.1 were included in the normal blood glucose group (NGR). Among the 55 participants who were included in the high blood glucose group (HGR), 22 had fasting plasma glucose level between 6.1 and 7.0 and were categorized as the impaired fasting glucose group (IFG), while 33 had fasting plasma glucose level no less than 7.0 and were categorized as the diabetic group (DM). Except for the fasting plasma glucose level, all other variables were comparable between NGR and HGR, or between NGR, IGF, and DM groups.
Table 1.
Demographic, biochemical and behavior characteristics of the participants
| Two Categorical | Three Categorical | Overall | |||||
|---|---|---|---|---|---|---|---|
| GLU normal (n = 78) | GLU high (n = 55) | NGR(n = 78) | IFG(n = 22) | DM(n = 33) | |||
| Numerical Variables | |||||||
| Age (year) | 69.08 ± 10.14 | 69.22 ± 10.04 | 69.08 ± 10.14 | 67.55 ± 10.80 | 70.33 ± 9.51 | 69.14 ± 10.06 | |
| BMI (kg/m2) | 25.5 ± 3.31 | 26.25 ± 3.02 | 25.5 ± 3.31 | 26.35 ± 3.04 | 26.18 ± 3.05 | 25.82 ± 3.2 | |
| WHR | 0.85 ± 0.07 | 0.88 ± 0.07 | 0.85 ± 0.07 | 0.87 ± 0.07 | 0.88 ± 0.07 | 0.87 ± 0.07 | |
| GLU (mmol/L) | 5.37 ± 0.42 | 7.76 ± 1.66 | 5.37 ± 0.42 | 6.47 ± 0.23 | 8.62 ± 1.65 | 6.36 ± 1.62 | |
| CHO (mmol/L) | 5.21 ± 1.05 | 5.17 ± 1.15 | 5.21 ± 1.05 | 5.04 ± 1.22 | 5.25 ± 1.10 | 5.19 ± 1.08 | |
| HDLC (mmol/L) | 1.48 ± 0.38 | 1.36 ± 0.27 | 1.48 ± 0.38 | 1.36 ± 0.23 | 1.36 ± 0.30 | 1.43 ± 0.34 | |
| LDLC (mmol/L) | 3.23 ± 0.78 | 3.31 ± 0.99 | 3.23 ± 0.78 | 3.17 ± 1.08 | 3.4 ± 0.94 | 3.26 ± 0.87 | |
| TG (mmol/L) | 1.91 ± 1.12 | 1.9 ± 1.38 | 1.91 ± 1.12 | 2.03 ± 1.70 | 1.82 ± 1.14 | 1.91 ± 1.23 | |
| SBP (mmHg) | 80.77 ± 10.95 | 78.16 ± 10.07 | 80.77 ± 10.95 | 79.23 ± 11.32 | 77.45 ± 9.26 | 79.69 ± 10.63 | |
| DBP (mmHg) | 140.24 ± 20.17 | 135.64 ± 17.22 | 140.24 ± 20.17 | 140.55 ± 20.63 | 132.36 ± 13.91 | 138.34 ± 19.08 | |
| Categorical Variables | |||||||
| Gender(n = 133) | male | 27 | 28 | 27 | 10 | 18 | 55 |
| female | 51 | 27 | 51 | 12 | 15 | 78 | |
| Edu3 (n = 123) | lower than college | 25 | 15 | 25 | 8 | 7 | 40 |
| college or higher | 46 | 37 | 46 | 13 | 24 | 83 | |
| Exercise (n = 99) | more than once a week | 38 | 34 | 38 | 12 | 22 | 72 |
| less than once a week | 16 | 11 | 16 | 6 | 5 | 27 | |
| Meat (n = 128) | not prefer | 37 | 21 | 37 | 10 | 11 | 58 |
| normal or prefer | 38 | 32 | 38 | 12 | 20 | 70 | |
| Fruit (n = 127) | not prefer | 27 | 23 | 27 | 9 | 14 | 50 |
| normal or prefer | 47 | 30 | 47 | 13 | 17 | 77 | |
| Vegetable (n = 126) | not prefer | 9 | 4 | 9 | 1 | 3 | 13 |
| normal or prefer | 64 | 49 | 64 | 21 | 28 | 113 | |
| Alcohol (n = 127) | more than 4 days a week | 3 | 3 | 3 | 2 | 1 | 6 |
| 1–3 days a week | 2 | 3 | 2 | 2 | 1 | 5 | |
| less than ones a week | 69 | 47 | 69 | 18 | 29 | 116 | |
| Smoke (n = 121) | not exposed | 49 | 29 | 49 | 13 | 16 | 78 |
| exposed 4 or more days a week | 13 | 20 | 13 | 8 | 12 | 33 | |
| exposed 1–3 days a week | 6 | 4 | 6 | 1 | 3 | 10 | |
| Sleeping (n = 124) | less than 6 h a day | 18 | 18 | 18 | 7 | 11 | 36 |
| more than 6 h a day | 55 | 33 | 55 | 13 | 20 | 88 | |
| Sleepless (n = 115) | more than 3 days a week | 8 | 5 | 8 | 1 | 4 | 13 |
| 1–3 days a week | 7 | 10 | 7 | 5 | 5 | 17 | |
| less than once a week | 51 | 34 | 51 | 13 | 21 | 85 | |
| Stress (n = 131) | happy | 44 | 33 | 44 | 13 | 20 | 77 |
| normal | 29 | 18 | 29 | 7 | 11 | 47 | |
| unhappy | 3 | 4 | 3 | 2 | 2 | 7 | |
Overall assessment of intestinal microbiota
A total of 25,375 K PE-reads of the 16S rDNA gene V3–V4 region were generated from the 133 specimens, with an average of 19,0791.3 (±39,560.7 SD) reads for each specimen, ranging from 85,363 to 368,127. A total of 19,305 K high quality PE-reads were obtained after trimming and filtering. In the OTU clustering process, a total of 37,668 sequences of chimeras were filtered and 7033 OTUs were yielded. After alignment of the OTU representative sequences using the QIIME pipeline, a total of 1366 OTUs were included for further data analysis.
In the taxonomical assignment process with a confidence threshold of 80%, 1366 operational taxonomic units (OTUs) were identified and annotated. Among these, 1302 OTUs at the phylum level had an annotation reliability over 0.8 and covered 16 phyla; 1231 OTUs at the class level had an annotation reliability over 0.8 and covered 28 classes; 1206 OTUs at the order level had an annotation reliability over 0.8 and covered 45 orders; 1056 OTUs at the family level had an annotation reliability over 0.8 and covered 99 families; and 652 OTUs at the genus level had an annotation reliability over 0.8 and covered 232 genera.
Firmicutes (47.9%) and Bacteroidetes (37.8) were found to be the dominant taxa at the phylum level, as were Clostridia (38.8%) and Bacteroidia (37.8%) at the class level and Clostridiales (38.8%) and Bacteroidales (37.8%) at the order level. At the family level, Bacteroidaceae (27.3%), Lachnospiraceae (21.0%), and Ruminococcaceae (15.6%) were the most dominant taxa covering more than 10% of the overall reads. At the genus level, a total of 53 genera had an abundance of more than 0.1%; among them, Bacteroides (27.3%), Prevotella (6.6%), Faecalibacterium (6.6%), Roseburia (5.2%), and Escherichia/Shigella (5.1%) were the most dominant taxa covering more than 5% of the overall reads.
Explanatory variables analysis
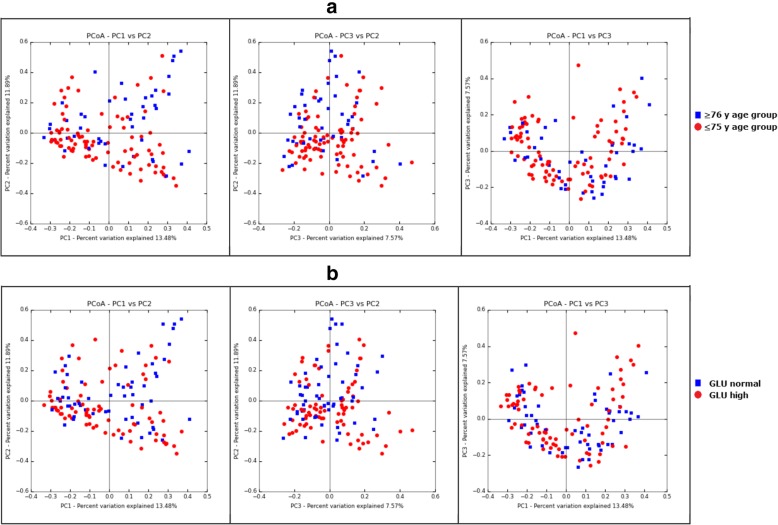
To understand the effects of subjects’ demographical, clinical, and behavioral characteristics on gut microbiota, we performed db-RDA analysis of each variable using Bray-Curtis matrix. The result shows that only factor of age and blood glucose level (GLU) have significant impacts on the Bray-Curtis distance matrix (Table 2). Further testing of the partial db-RDA analysis, which allows the influence of a matrix of conditioning variables to be partialed-out prior to analysis and thus enables evaluation of independent impacts of each individual factor on the gut microbiota structure shows that both GLU and age had a significant independent impact on the Bray-Curtis distance. Notably, the age factor has the most significant impact on the Bray-Curtis distance matrix, which explained 0.9% of total variances, independently. The 2D PCoA plots of the stool microbiota of people from two age group and different glucose levels based on Bray-Curtis distance matrix was shown in Fig. 1.
Table 2.
Analysis of the association of demographic, biochemical and behavior characteristics variants and the microbiota based on db-RDA and Partial db-RDA on Bray-Curtis distance matrix
| db-RDA | Partial db-RDA | |||
|---|---|---|---|---|
| Adjusted R2 | p | Adjusted R2 | p | |
| Gender | 0.000786 | 0.286 | ||
| Age | 0.009047 | 0.003 | 0.00905 | 0.001 |
| Edu3 | 0.000428 | 0.362 | ||
| Exercise | 0.001449 | 0.248 | ||
| Meat | −0.00074 | 0.583 | ||
| Fruit | −0.00051 | 0.534 | ||
| Vegetable | 0.001639 | 0.214 | ||
| Alcohol | −0.0008 | 0.54 | ||
| Smoke | 0.001909 | 0.249 | ||
| Sleeping | −0.00198 | 0.809 | ||
| Sleepless | 0.001241 | 0.293 | ||
| Stress | −0.00141 | 0.668 | ||
| SBP | 0.001487 | 0.181 | ||
| DBP | −0.00023 | 0.494 | ||
| BMI | −0.00147 | 0.834 | ||
| WHR | 0.000624 | 0.281 | ||
| GLU | 0.004232 | 0.017 | 0.00423 | 0.01 |
| CHO | 0.003007 | 0.057 | ||
| HDLC | 0.002124 | 0.11 | ||
| LDLC | 0.000757 | 0.272 | ||
| TG | −0.00045 | 0.561 | ||
Boldface indicate P< 0.05
Fig. 1.
The 2D PCoA plots of the stool microbiota of people from two age group and different glucose levels based on Bray-Curtis distance matrix. (a) Comparison between the ≥ 76 y age group and ≤ 75 y age group. (b) Comparison between the GLU normal group and GLU high group
Age supervised clustering of the microbiota
To investigate the impact of the age on the gut microbiota composition, we analyzed bacterial differentiated abundance using age-supervised multivariate regress trees (MRT). The results showed that when supervised by age, the microbiota could be stratified into two groups with 75.5 years as the cut-off age (Fig. 2).
Fig. 2.
Clustering of the microbiota data supervised by the age based on MRT (multivariate regression trees) analysis and the result of variation partitioning analysis exploring explanatory power of numerical and categorial variant of age in relation to the microbiota data matrix. (a) The MRT analysis indicates that supervised by the age variant, the microbiota data could be stratified as 75 and younger group and 76 and older group. (b) The variation partitioning analysis indicates that the most part of the explanatory power of the numerical variant of age in relation to the microbiota data matrix could be explained by the two categorial variant of age (age of ≤75 or ≥ 76)
Variation partitioning analysis was conducted to explore the explanatory power of a numerical age variant or a categorical age variant. The results indicated that using a categorical age variant with 75.5 years as the cut-off value could explain the majority of age-related gut microbiota changes.
Comparison of diversity of microbial communities among groups with different glucose levels and age levels
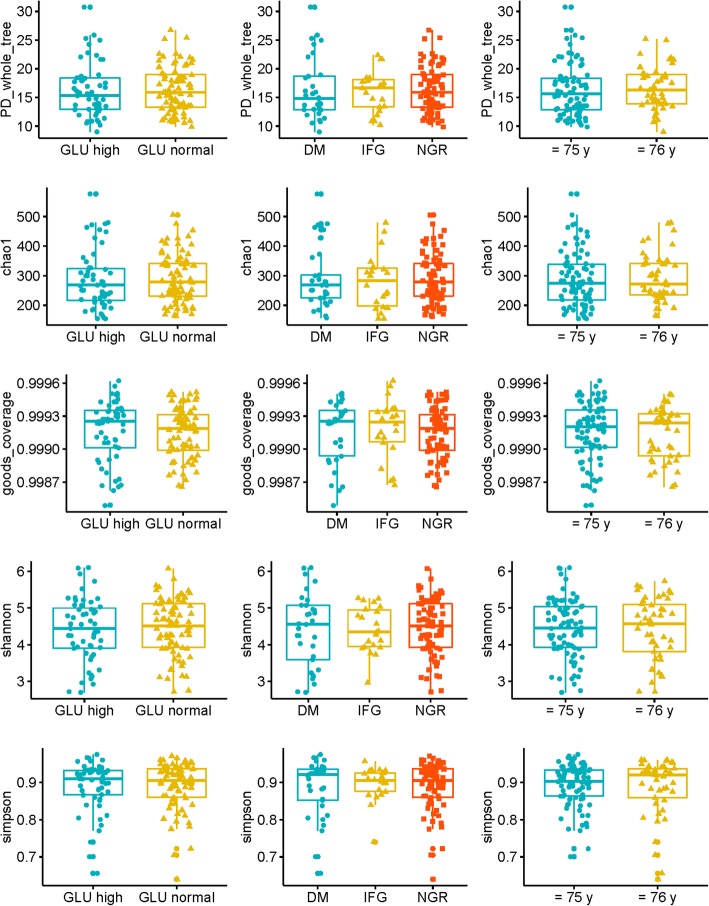
To compare the alpha diversity, the Chao1, Shannon, Simpson, PD whole tree, and Good’s coverage indexes were calculated after randomly subsampling the OTU table down to 60,265 reads per sample, the size of the smallest sample to obtain equal sequencing depth. No significant differences were found between different groups of glucose levels and age levels (age of ≤75 y or ≥ 76 y) (Table 3, Fig. 3).
Table 3.
Comparison of the alpha diversity indexes across different group of samples
| Overall comparison | ||||||||||||
| Two Categorical GLU group | Three Categorical GLU group | Age group | Total (n = 133) | |||||||||
| GLU normal (n = 78) | GLU high (n = 55) | NGR (n = 78) | IFG (n = 22) | DM (n = 33) | 76 or older (n = 45) | 75 or younger (n = 88) | min | median | max | mean | sd | |
| PD_whole_tree | 16.4 ± 4 | 16.2 ± 4.6 | 16.4 ± 4 | 16 ± 3.5 | 16.3 ± 5.3 | 16.6 ± 3.8 | 16.2 ± 4.5 | 9 | 15.8 | 30.7 | 16.3 | 4.2 |
| Chao1 | 290.3 ± 82.7 | 286.2 ± 98.3 | 290.3 ± 82.7 | 278.6 ± 92 | 291.3 ± 103.4 | 291.4 ± 81.9 | 287.2 ± 93 | 154.5 | 272.2 | 576.5 | 288.6 | 89.1 |
| Goods_coverage | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 | 1 | 1 | 1 | 0 |
| Shannon | 4.5 ± 0.8 | 4.4 ± 0.9 | 4.5 ± 0.8 | 4.4 ± 0.6 | 4.4 ± 1 | 4.4 ± 0.9 | 4.5 ± 0.8 | 2.7 | 4.5 | 6.1 | 4.4 | 0.8 |
| Simpson | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0 | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.6 | 0.9 | 1 | 0.9 | 0.1 |
| Comparison among 76 ages and older subgroup | ||||||||||||
| Two Categorical GLU group | Three Categorical GLU group | Total (n = 45) | ||||||||||
| GLU normal (n = 26) | GLU abnormal (n = 19) | NGR (n = 26) | IFG (n = 6) | DM (n = 13) | min | median | max | mean | sd | |||
| PD_whole_tree | 17.2 ± 3.9 | 15.8 ± 3.7 | 17.2 ± 3.9 | 15.8 ± 3.7 | 15.8 ± 3.9 | 9.0 | 16.3 | 25.2 | 16.6 | 3.8 | ||
| Chao1 | 301.3 ± 81.9 | 278 ± 82.1 | 301.3 ± 81.9 | 262.7 ± 110.6 | 285 ± 69.7 | 162.3 | 272.1 | 479.5 | 291.4 | 81.9 | ||
| Goods_coverage | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | ||
| Shannon | 4.6 ± 0.8 | 4.2 ± 0.9 | 4.6 ± 0.8 | 4.2 ± 0.8 | 4.2 ± 1 | 2.7 | 4.6 | 5.7 | 4.4 | 0.9 | ||
| Simpson | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.6 | 0.9 | 1.0 | 0.9 | 0.1 | ||
| Comparison among 75 ages and younger subgroup | ||||||||||||
| Two Categorical GLU group | Three Categorical GLU group | Total (n = 88) | ||||||||||
| GLU normal (n = 52) | GLU abnormal (n = 36) | NGR (n = 52) | IFG (n = 16) | DM (n = 20) | min | median | max | mean | sd | |||
| PD_whole_tree | 16 ± 4 | 16.4 ± 5.1 | 16 ± 4 | 16.1 ± 3.6 | 16.6 ± 6.1 | 9.9 | 15.6 | 30.7 | 16.2 | 4.5 | ||
| Chao1 | 284.8 ± 83.3 | 290.6 ± 106.7 | 284.8 ± 83.3 | 284.6 ± 87.3 | 295.4 ± 122 | 154.5 | 274.2 | 576.5 | 287.2 | 93.0 | ||
| Goods_coverage | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1 ± 0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | ||
| Shannon | 4.4 ± 0.7 | 4.5 ± 0.8 | 4.4 ± 0.7 | 4.5 ± 0.5 | 4.5 ± 1 | 2.7 | 4.5 | 6.1 | 4.4 | 0.8 | ||
| Simpson | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0 | 0.9 ± 0.1 | 0.7 | 0.9 | 1.0 | 0.9 | 0.1 | ||
Fig. 3.
Comparison the alpha diversity between different groups of glucose levels and age levels
In the analyses of beta diversity, consistent with the result of db-RDA analysis, Adonis tests also shows that both glucose level variance and age levels (age of ≤75 y or ≥ 76 y) were significantly associated with Bray-Curtis distance matrix. However, in the Adonis test after stratification into ≤75 y and ≥ 76 y age groups, the association between glucose levels and Bray-Curtis distance matrix for the≤75 y age group was no longer significant, while in the ≥76 y age group, the association was still significant with a significant higher R-square values (Table 4). These results suggested that the gut microbiota structure differed more significantly as glucose levels changed in the ≥76 y age group than in the ≤75 y age group.
Table 4.
Adonis analysis on the association of microbiota and different glucose levels based on Bray-Curtis distance matrixes
| GLU normal VS GLU high | NGR VS IGF VS DM | |||
|---|---|---|---|---|
| R square value | P value | R square value | P value | |
| Overall data | 0.01961 | 0.003 | 0.05439 | 0.002 |
| Aged 76 and older group | 0.09618 | 0.002 | 0.11801 | 0.002 |
| Aged 75 and younger group | 0.01955 | 0.117 | 0.03661 | 0.091 |
Boldface indicate P< 0.05
We compared the microbiota Bray-Curtis distance matrix of groups with different ages and glucose levels using the Wilcoxon signed-rank test. The results showed that although inter-individual variation within groups was considerably high, that of the low glucose group was still significantly lower than that of the high glucose group, and that of the ≤75 y age group was significantly lower than that of the ≥76 y age group. After stratification into ≤75 y and ≥ 76 y age groups, the inter-individual variation of the high glucose group was significantly higher than that of the normal glucose group (Fig. 4).
Fig. 4.
Comparison of inter-individual variation within group among samples from different groups of glucose levels stratified by age
Differences in stool microbiota taxa between groups with different glucose levels
In Wilcoxon rank-sum test, a total of 9 taxa were found to have different abundances between the GLU normal group and GLU high group (q < 0.05 after correction for multiple testing by false discovery rate (FDR) control with the Benjamini–Hochberg procedure) (Table 5). Among them, the Actinobacteria at phylum level, Actinobacteria and Bacilli at class level, Bifidobacteriales, and Lactobacillales at order level, Bifidobacteriaceae and Lactobacillaceae at family level, and Lactobacillus and Bifidobacterium at genus level were more abundant in the GLU high group.
Table 5.
Differences in stool microbiota taxon between different groups of glucose levels
| Overall comparison | ||||||||||||
| Taxon |
GLU normal (Mean ± SD) |
GLU high (Mean ± SD) |
Wilcoxon rank-sum test | Discriminatory power |
GLU normal (Mean ± SD) |
DM (Mean ± SD) |
Wilcoxon rank-sum test | Discriminatory power | ||||
| p-value | q-value | AUC value | sig. | p-value | q-value | AUC value | sig. | |||||
| p__Actinobacteria | 0.0253 ± 0.0363 | 0.0761 ± 0.0921 | 0.0003 | 0.018 | 0.6848 | 0.0004 | 0.0253 ± 0.0363 | 0.0846 ± 0.088 | 0 | 0.0028 | 0.7463 | 0.0003 |
| p__Actinobacteria.c__Actinobacteria | 0.0253 ± 0.0363 | 0.0761 ± 0.0921 | 0.0003 | 0.018 | 0.6848 | 0.0004 | 0.0253 ± 0.0363 | 0.0846 ± 0.088 | 0 | 0.0028 | 0.7463 | 0.0003 |
| p__Actinobacteria.c__Actinobacteria.o__Bifidobacteriales | 0.0161 ± 0.026 | 0.0631 ± 0.0815 | 0.0001 | 0.0099 | 0.6976 | 0.0003 | 0.0161 ± 0.026 | 0.0714 ± 0.0855 | 0 | 0.0028 | 0.7475 | 0.0004 |
| p__Actinobacteria.c__Actinobacteria.o__Bifidobacteriales.f__Bifidobacteriaceae | 0.0161 ± 0.026 | 0.0631 ± 0.0815 | 0.0001 | 0.0099 | 0.6976 | 0.0003 | 0.0161 ± 0.026 | 0.0714 ± 0.0855 | 0 | 0.0028 | 0.7475 | 0.0004 |
| p__Actinobacteria.c__Actinobacteria.o__Bifidobacteriales.f__Bifidobacteriaceae.g__Bifidobacterium | 0.0161 ± 0.026 | 0.062 ± 0.0803 | 0.0001 | 0.0099 | 0.6969 | 0.0003 | 0.0161 ± 0.026 | 0.0696 ± 0.084 | 0 | 0.0028 | 0.7457 | 0.0004 |
| p__Firmicutes.c__Bacilli | 0.0033 ± 0.0069 | 0.0357 ± 0.0942 | 0.0008 | 0.042 | 0.6714 | 0.0021 | 0.0033 ± 0.0069 | 0.0315 ± 0.0799 | 0.001 | 0.0476 | 0.6981 | 0.0028 |
| p__Firmicutes.c__Bacilli.o__Lactobacillales | 0.0032 ± 0.0069 | 0.0353 ± 0.0923 | 0.0009 | 0.0444 | 0.669 | 0.0021 | 0.0032 ± 0.0069 | 0.0315 ± 0.0799 | 0.001 | 0.0476 | 0.6985 | 0.0027 |
| p__Firmicutes.c__Bacilli.o__Lactobacillales.f__Lactobacillaceae | 0.0011 ± 0.0053 | 0.0271 ± 0.0812 | 0 | 0.0099 | 0.7064 | 0.0155 | 0.0011 ± 0.0053 | 0.0238 ± 0.0752 | 0 | 0.0028 | 0.7479 | 0.0268 |
| p__Firmicutes.c__Bacilli.o__Lactobacillales.f__Lactobacillaceae.g__Lactobacillus | 0.0011 ± 0.0053 | 0.0271 ± 0.0812 | 0 | 0.0099 | 0.7064 | 0.0155 | 0.0011 ± 0.0053 | 0.0238 ± 0.0752 | 0 | 0.0028 | 0.7479 | 0.0268 |
| Comparison among 76 ages and older subgroup | ||||||||||||
| Taxon |
GLU normal (Mean ± SD) |
GLU high (Mean ± SD) |
Wilcoxon rank-sum test | Discriminatory power |
GLU normal (Mean ± SD) |
DM (Mean ± SD) |
Wilcoxon rank-sum test | Discriminatory power | ||||
| p-value | q-value | AUC value | sig. | p-value | q-value | AUC value | sig. | |||||
| p__Actinobacteria | 0.0293 ± 0.0363 | 0.1322 ± 0.0917 | 0 | 0.0004 | 0.8725 | 0.0011 | 0.0293 ± 0.0363 | 0.1581 ± 0.0934 | 0 | 0.0002 | 0.926 | 0.0017 |
| p__Actinobacteria.c__Actinobacteria | 0.0293 ± 0.0363 | 0.1322 ± 0.0917 | 0 | 0.0004 | 0.8725 | 0.0011 | 0.0293 ± 0.0363 | 0.1581 ± 0.0934 | 0 | 0.0002 | 0.926 | 0.0017 |
| p__Actinobacteria.c__Actinobacteria.o__Bifidobacteriales | 0.0231 ± 0.0348 | 0.1169 ± 0.0907 | 0 | 0.0004 | 0.8725 | 0.0015 | 0.0231 ± 0.0348 | 0.1384 ± 0.0959 | 0 | 0.0002 | 0.9201 | 0.0023 |
| p__Actinobacteria.c__Actinobacteria.o__Bifidobacteriales.f__Bifidobacteriaceae | 0.0231 ± 0.0348 | 0.1169 ± 0.0907 | 0 | 0.0004 | 0.8725 | 0.0015 | 0.0231 ± 0.0348 | 0.1384 ± 0.0959 | 0 | 0.0002 | 0.9201 | 0.0023 |
| p__Actinobacteria.c__Actinobacteria.o__Bifidobacteriales.f__Bifidobacteriaceae.g__Bifidobacterium | 0.0231 ± 0.0348 | 0.1164 ± 0.09 | 0 | 0.0004 | 0.8745 | 0.0015 | 0.0231 ± 0.0348 | 0.1378 ± 0.0951 | 0 | 0.0002 | 0.9201 | 0.0022 |
| p__Bacteroidetes.c__Bacteroidia.o__Bacteroidales.f__Bacteroidaceae | 0.29 ± 0.1638 | 0.1333 ± 0.1286 | 0.0015 | 0.0464 | 0.7733 | 0.0052 | 0.29 ± 0.1638 | 0.1249 ± 0.1186 | 0.0028 | 0.0867 | 0.7899 | 0.0094 |
| p__Bacteroidetes.c__Bacteroidia.o__Bacteroidales.f__Bacteroidaceae.g__Bacteroides | 0.29 ± 0.1638 | 0.1333 ± 0.1286 | 0.0015 | 0.0464 | 0.7733 | 0.0052 | 0.29 ± 0.1638 | 0.1249 ± 0.1186 | 0.0028 | 0.0867 | 0.7899 | 0.0094 |
| p__Firmicutes.c__Bacilli | 0.0032 ± 0.0053 | 0.0822 ± 0.1438 | 0.0001 | 0.0042 | 0.834 | 0.0047 | 0.0032 ± 0.0053 | 0.068 ± 0.1172 | 0.0002 | 0.0091 | 0.8491 | 0.0048 |
| p__Firmicutes.c__Bacilli.o__Lactobacillales | 0.0032 ± 0.0053 | 0.0811 ± 0.1406 | 0.0001 | 0.0042 | 0.832 | 0.0047 | 0.0032 ± 0.0053 | 0.0679 ± 0.1171 | 0.0002 | 0.0091 | 0.8491 | 0.0048 |
| p__Firmicutes.c__Bacilli.o__Lactobacillales.f__Lactobacillaceae | 0.0012 ± 0.0037 | 0.0647 ± 0.1244 | 0 | 0.0001 | 0.9069 | 0.0259 | 0.0012 ± 0.0037 | 0.0574 ± 0.1116 | 0 | 0.0002 | 0.9201 | 0.0262 |
| p__Firmicutes.c__Bacilli.o__Lactobacillales.f__Lactobacillaceae.g__Lactobacillus | 0.0012 ± 0.0037 | 0.0647 ± 0.1244 | 0 | 0.0001 | 0.9069 | 0.0259 | 0.0012 ± 0.0037 | 0.0574 ± 0.1116 | 0 | 0.0002 | 0.9201 | 0.0262 |
| p__Firmicutes.c__Clostridia.o__Clostridiales.f__Lachnospiraceae | 0.233 ± 0.12 | 0.1087 ± 0.0799 | 0.0003 | 0.0153 | 0.8057 | 0.0031 | 0.233 ± 0.12 | 0.0925 ± 0.0574 | 0.0002 | 0.0091 | 0.855 | 0.0048 |
| p__Firmicutes.c__Clostridia.o__Clostridiales.f__Lachnospiraceae.g__Lachnospiracea_incertae_sedis | 0.0305 ± 0.026 | 0.0107 ± 0.0099 | 0.0014 | 0.0464 | 0.7753 | 0.0147 | 0.0305 ± 0.026 | 0.01 ± 0.0094 | 0.0028 | 0.0867 | 0.7899 | 0.0303 |
The discriminatory power of taxa was further assessed by calculating the AUC of a logistic regression model. As a result, the AUC values of all 9 taxa were above 0.65, and among them, the taxa Lactobacillus at genus level and Lactobacillaceae at family level were shown to have AUC values higher than 0.7, which represents reasonable discrimination power.
The HGR was further divided into IFG and DM subgroups, which were then compared with the NGR group. No taxa were found have different abundances between the NGR group and IFG group, after correction for multiple testing by FDR control with the Benjamini–Hochberg procedure. In the comparison of the NGR group and DM group, all the 9 taxa, which were more abundant in the GLU high group comparing to GLU normal group were also found have different abundances (q < 0.05 after correction for multiple testing by FDR control with the Benjamini–Hochberg procedure). The AUC values of the 9 taxa ranged from 0.69 to 0.75, and the AUC values of taxa Actinobacteria at phylum level, Actinobacteria at class level, Bifidobacteriales at order level, Bifidobacteriaceae and Lactobacillaceae at family level, and Bifidobacterium and Lactobacillus at genus level were higher than 0.7, which represents reasonable discrimination power.
Comparison of stool microbiota taxa between groups with different glucose levels after age-stratification using 76 years as the cut-off age
We compared differentiated taxa in groups with different glucose levels after stratification with 75-years as the cut-off age using the Wilcoxon rank-sum test (Table 5). The results showed no differentiated taxa between groups with different glucose levels in the ≤75 y age group. However, in the ≥76 y age group, 13 taxa showed differentiated abundances between the high GLU group and normal GLU group (q < 0.05 after correction for multiple testing by FDR control with the Benjamini–Hochberg procedure). The Bacteroidaceae and Lachnospiraceae at family level, and Lachnospiraceae incertae sedis and Bacteroides at genus level were more abundant in the GLU normal group, whereas the Actinobacteria at phylum level, Actinobacteria and Bacilli at class level, Bifidobacteriales and Lactobacillales at order level, Lactobacillaceae and Bifidobacteriaceae at family level, and Lactobacillus and Bifidobacterium at genus level were more abundant in the GLU high group. All had AUC values greater than 0.80. Among the 13 taxa, Lactobacillaceae and Lactobacillus had AUC values greater than 0.90, which suggested these two taxa had satisfactory discrimination power. Among those 13 taxa, 10 taxa also showed differentiated abundances between NGR and DM groups with AUC values greater than 0.85. no taxa showed differentiated abundances between the NGR and IFG group.
Discussion
In this study, we investigated the association between gut microbiota and variables including plasma glucose levels and demographic, behavioral, and biochemical characteristics in a population with chronic disease. The results indicated that plasma glucose level and age contributed significantly to a differentiated gut microbiota structure. We also identified several taxa whose abundance differed significantly in subgroups with different glucose levels. After age stratification, we found that the plasma glucose level affected the gut microbiota structure more significantly in the ≥76 y age group than in the ≤75 y age group. Moreover, the taxon abundance changed significantly with glucose level in different age groups.
With the widespread application of next-generation sequencing technology, more and more studies have demonstrated a close relationship between gut microbiota and human health and disease. Multiple host demographic factors and behavioral factors play important and confounding roles in the relationship between gut microbiota and physio-pathological indicators.
Our study analyzed the association between gut microbiota structure and glucose level and multiple demographic, behavioral, and biochemical factors using db-RDA. The results demonstrated that age and glucose level were significantly associated with gut microbiota structure.
Several studies have investigated the relationship between age or glucose level and microbiota composition, and several hypotheses have been proposed. During the aging process, the physiology of the intestinal tract is affected, dietary habits and lifestyles change, and immunosenescence occurs, all of which contribute to age-related imbalance of the intestinal microbial community. Reported age-related changes in the intestinal microbiota include dysbiosis, loss of microbial diversity, increased vulnerability to environmental perturbations, loss of probiotics, shifts in the dominant species within several bacterial groups, increase in the total number of facultative anaerobes, and reduced SCFA production rates. These modifications of the intestinal microbiota may contribute to risk for several diseases like inflammatory bowel conditions, metabolic diseases, as well as musculoskeletal conditions [24]. Previous studies have suggested that abnormal blood glucose levels might cause gut microbiota changes, such as dysbacteriosis, compositional changes of microbiota, and changes in metabolites. For example, as the blood glucose level changes, the concentration of short-chain fatty acids (SCFAs), which have significant immune system effects in the intestinal mucosa, decreases, while the concentration of LPS from gram-negative bacteria increases. As a result, the pro-inflammatory signal transduction pathway is activated and consequently causes chronic low-grade inflammatory status, reduced insulin sensitivity, and a series of changes that eventually lead to the occurrence of T2D [25, 26]. All of these studies indicate that dysbacteriosis is closed associated with aging and abnormal blood glucose levels [25, 26]. However, an objective and comprehensive definition of intestinal flora imbalances is still lacking.
Some studies have attempted to identify the alpha-diversity related characteristics in the gut microbiota of aged populations. Biage et al. observed that the microbial composition and diversity of the gut ecosystem of young adults differs significantly from that of Italian centenarians [27]. However, Bian et al. and Kong et al. reported that the microbiota of healthy aged adults differs little from that of healthy young adults in the Chinese population [28, 29]. Similarly, it is debatable whether alpha-diversity related microbiota characteristics are associated with abnormal blood glucose levels, with some studies reporting associations between lower microbiota diversity and T2D or insulin resistance [16, 30], while others do not support such associations [14, 17, 20]. In this study, we compared five alpha-diversity indices between different age groups with various blood glucose levels and did not find significant differences. Discrepancies in the association between microbiota alpha-diversity and age or blood glucose levels could be explained by ethnic or demographic difference between studies. For example, Wang et al. investigated and compared the composition and richness of the gut microbiota of healthy individuals and diabetes patients from two ethnic groups, Uyghurs and Kazaks. Significant differences in microbial richness and a higher number of OTUs were found between the Kazak healthy and diabetic groups, while no major differences in intestinal microbiota were found between the Uyghur healthy and diabetic groups [13]. Another possible explanation might be that alpha-diversity is affected by multiple confounding factors. When the factors are not well controlled, the results might be biased.
Inter-individual variation within groups is another parameter that can reflect the stability of gut microbiota. Individuals with healthy and hemostatic gut microbiota share higher similarity regarding the composition and richness of microbes, while those with imbalanced microbiota show different changes and tend to have higher inter-individual variations. This is similar to the so-called Anna Karenina principle, derived from Leo Tolstoy’s dictum that “all happy families look alike; each unhappy family is unhappy in its own way”, and has been used for successful modeling in many different fields such as business, psychology, economics, biology, and recently in microbiota [31]. In this study, we compared the inter-group differences of the distances matrix under different age and plasma glucose conditions using the Wilcoxon signed-rank test. The results showed considerable inter-individual variation within groups. However, the inter-individual variation of the low blood glucose group was significantly lower than that of the high blood glucose group, and the inter-individual variation of the ≤75 y age group was significantly lower than that of the ≥76 y age group. These results are consistent with Bian’s report [28] in which the 94-year-old group had a larger beta diversity than did younger groups, and with Qin’s report [14] in which T2D was found to be a significant factor in the variation in examined gut microbial samples. These studies all support the hypothesis that microbiota homeostatic imbalance is age and blood sugar level-related. The elderly or individuals with abnormal blood sugar levels tend to have higher inter-individual variation in gut microbes or the variation tends to be greater.
Further Wilcoxon signed-rank tests in the age-stratified groups showed that although the inter-individual variation of the high-blood glucose group was significantly higher than that of NGP in both ≤75 y and ≥ 76 y age groups, compared with the ≤75 y age group, the differences of the inter-individual variation were more significant in the ≥76 y age group. This result is consistent with the Adonis analysis of the age-stratified groups and indicated that the association between blood glucose level and gut microbiota stability differs in different age groups. Therefore, it is necessary to consider the effects of age when investigating the relationship between blood glucose and gut microbiota.
In this study, we analyzed the taxonomic differences in gut microbiota in groups with different blood glucose levels. We found that Lactobacillus species and their related taxa had higher richness in the high blood glucose group (> 6.0) as compared with the normal blood glucose group (≤6.0). Similarly, Lactobacillus species and their related taxa had higher richness in the DM group than in the NGT group. This is consistent with previous studies on diabetic subjects in different populations of the world indicating a significantly higher abundance of Lactobacillus species in fecal samples of high blood glucose groups [16, 19, 20]. In children with insulin-dependent diabetes mellitus (IDDM), high salivary glucose levels lead to increased salivary lactobacilli counts [32]. Therefore, increased abundance of Lactobacillus in the gastrointestinal tract could be the result of increased intestinal glucose levels [16]. In fact, our results also demonstrated that the abundance of Lactobacillus tends to increase as the blood glucose level increases in NGT, IFG, and DM groups.
The richness of Bifidobacterium species and their related taxa was also found significantly associated with elevated blood glucose levels. This result is contrary to those of Wu et al. [18] and Sedighi et al. [19]. They found lower concentrations of Bifidobacterium in T2DM patients than in normal controls. However, our result is consistent with that of Sepp et al., in which the counts and proportions of Bifidobacterium were associated with higher glucose levels [15]. Therefore, the association between the richness of Bifidobacterium species and blood glucose levels is controversial and needs further investigation.
The controversy over the association between gut microflora composition and blood glucose levels could be explained by differences in the participating populations, the microbial detection methods, the statistical methods, etc. In addition, multiple confounding factors could cause contradictory results. In our study, we can conclude that age is an important factor affecting the association between blood glucose levels and gut microbial composition.
It is generally accepted that an age of 60 or 65 years is defined as elderly or old. However, the definition of aged gut microbiota is still debatable. Current investigations of the association between age and gut microbiota are mainly based on different age groups, but the results vary significantly [27, 28, 33, 34]. When conducting age-related microbiota studies, using study-specific age standard may improve the reliability of the results. Therefore, we used a supervised clustering method and found the microbiota could be strictly stratified into two groups with 75.5 years as the cut-off age. Further variation partitioning analysis shows that the categorical age variant with 75.5 years as the cut-off value could explain the majority of age-related gut microbiota changes, therefore the age of 75.5 years might be the best cut-off to stratify the population of this study. The finding of the potential switch in the microbiota structure at the age of 76 years in this study population is very interesting and indicative, however more researches must be conducted to evaluate whether the cut off age of 75.5 years is also applicable to other populations.
After stratification by age at 75 years, the differentiated taxa were analyzed and compared in groups with different blood glucose levels. The result showed that the blood glucose-related taxa differed significantly between the ≤75 y and ≥ 76 y age groups. In the ≤75 y age group, the differences of blood glucose-related taxa showed a similar trend as in the ≥76 y age group. However, after correcting by q value, the difference was not significant (data not shown). In the ≥76 y age group, the differences of these taxa were much more significant. For example, Lactobacillus genus and 3 related taxa, as well as Bifidobacterium genus and 5 related taxa, were able to differentiate high glucose and normal glucose, as well as DM and NGT, with an AUC value greater than 0.8.
In addition, in the ≥76 y age group, 2 taxa, the Lachnospiraceae family and the Lachnospiracea incertae sedis genus showed significantly higher abundances in the normal blood glucose group. Of them, the Lachnospiraceae family also displayed significant associations with blood glucose level when we compared the DM and NGR groups. An association between the Lachnospiraceae family and blood glucose level has been reported, but the results are inconsistent. According to Bhute et al. [16], Lachnospiraceae were significantly more abundant in NGTs subjects than in the DM group, while Qin et al. reported that an metagenomic linkage group (MLG) assigned to the Lachnospiraceae family was significantly associated with T2D [14]. In our study, the association between Lachnospiraceae and its related taxa and blood glucose levels were not significant in the ≤75 y age group (p > 0.05), but very significant in ≥76 y age DM group with all the AUC values greater than 0.85 (p < 0.01, q > 0.05). These results suggested that the associations between these taxa and blood glucose levels might change with different age levels.
There are some limitations of this study. First, due to the cross-sectional experimental design, we were not able to determine the causal connection between aging, abnormal glucose, and changes in gut microbiota. Second, we used 16 sRNA gene sequencing to analyze the gut microbiome. This method could introduce bias during several processes, including the selection of the gene amplification area, the gene amplification procedure, the selection of gene sequence database, and OTU clustering. In addition, this study included many elderly participants. Older people generally have a higher burden of multimorbidity and polypharmacy than younger ones, and both these elements could be associated with a different fecal microbiota composition. This could also introduce some bias in the results of this study. However, compared with other studies that have investigated the relationship between blood glucose level and gut microbiota, the strengths of our work includes, we enrolled participants with a wider age range, all the participants were from the same community and their behavioral and physiological parameters were collected.
Conclusions
In this study, we found that both blood glucose level and age have significant impacts on the composition of gut microbiota. The association between glucose level and the composition and activity of gut microbiota was affected profoundly by age and displayed distinct characteristics at different age groups. Our findings suggest that it is necessary to take age into account when investigating the association between glucose and gut microbiota. We also identified multiple taxa that were highly associated with high glucose levels in the ≥76 y age group, but not in the ≤75 y age group. More research is required to determine the underlying biological mechanisms.
Methods
Sample collection and processing
With approval from the Ethics Committee of Minzu University of China (MUC), the subjects of the present study were enrolled from populations that participated in community health examinations in Beijing in August 2015. The inclusion criteria included 1) Subject is a male or female aged over 40. 2) Subject did not take any medicine in recent 2 weeks. 3) subject have no previous chronic gastrointestinal disease. The exclusive criteria included usage of any medicine, probiotics, or prebiotics within 2 weeks, diagnosis of psychiatric disorders, intestinal diseases, and neoplasia. A total of 133 subjects were enrolled in the study after obtaining both written and verbal consent from the subjects.
Height, weight, waist-to-hip ratio, and blood pressure were measured for all subjects, and risk factor information of lifestyle profile (such as exercise, diet, smoking, alcohol, sleeping, and stress) were collected by a questionnaire during the waiting time of the physical examination. Blood samples before breakfast were collected to measure blood lipids and glucose. Fecal samples were collected with sterile cups and were frozen at − 20 °C immediately, then transferred to the laboratory within 24 h and stored at − 80 °C before DNA extraction.
Blood samples before breakfast were tested for glucose, serum total cholesterol (CHO), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), and triglycerides (TG) using an automatic biochemical analyzer.
Extraction of total DNA from stool samples was conducted according to the manual of the PowerSoil® DNA Isolation Kit. The purity of DNA was evaluated using the A260/280 ratio. Samples with A260/280 ratios between 1.8 and 2.2 were used for further experiments.
Microbiota sequencing
The V3-V4 region of the bacterial 16S rRNA gene was amplified with the common primer pair (forward primer, 5′- ACTCCTACGGGAGGCAGCA-3′; reverse primer, 5′- GGACTACHVGGGTWTCTAAT-3′) combined with adapter sequences and barcode sequences. PCR amplification was performed in a total volume of 50 μl, which contained 10 μl buffer, 0.2 μl Q5 High-Fidelity DNA Polymerase, 10 μl High GC Enhancer, 1 μl dNTP, 10 μM of each primer, and 60 ng genomic DNA. Thermal cycling conditions were as follows: an initial denaturation at 95 °C for 5 min, followed by 15 cycles at 95 °C for 1 min, 50 °C for 1 min, and 72 °C for 1 min, with a final extension at 72 °C for 7 min. The PCR products from the first step PCR were purified through VAHTSTM DNA Clean Beads. A second round of PCR was then performed in a 40-μl reaction which contained 20 μl 2 × Phusion HF MM, 8 μl ddH2O, 10 μM of each primer, and 10 μl PCR products from the first step. Thermal cycling conditions were as follows: an initial denaturation at 98 °C for 30 s, followed by 10 cycles at 98 °C for 10 s, 65 °C for 30 s min, and 72 °C for 30 s, with a final extension at 72 °C for 5 min. Finally, all PCR products were quantified by the Quant-iT™ dsDNA HS Reagent and pooled together. High-throughput sequencing analysis of bacterial rRNA genes was performed on the purified, pooled sample using the Illumina Hiseq 2500 platform (2 × 250 paired ends) at Biomarker Technologies Corporation, Beijing, China.
Bioinformatics analysis
The raw reads were demultiplexed and then trimmed, merged, and filtered by Usearch9.0.2132_i86linux32 following the UPARSE pipeline. All reads were trimmed to the position of the first base with quality score ≤ 2, and sequences shorter than 64 after trimming were discarded. Paired reads with a number of expected error > 1.00 were further filtered out during the filtering step. Sequences were dereplicated and clustered with a threshold of 97% similarity for picking operational taxonomic units (OTUs) representative after Chimera checking. After that, all sequences were mapped back to the representative sequences resulting in an OTU table for all samples. The RDP Classifier was used to assign 16S rRNA gene sequences to a taxonomical hierarchy with a confidence threshold of 80%. OTU representative sequences were aligned and further filtered to create a phylogenetic tree using the QIIME pipeline. The OTU table was randomly subsampled down to the size of the smallest sample to obtain equal sequencing depth. Finally, a total of 60,265 reads per sample were used for further analysis.
Statistical analysis
Categorical variables are presented as frequencies and percentages. Chi-squared tests and Fisher’s exact test were used to assess statistical associations between variables. Numerical variables are expressed as mean ± standard deviation (SD). ANOVA (one-way analysis of variance) was used to compare the differences between groups.
The Chao1, Shannon, Simpson, PD whole tree, and Good’s coverage indexes were used for richness and diversity estimations of the gut microbiota.
The db-RDA and Adonis tests were performed on the Bray-Curtis distance matrix to investigate the differences of beta diversity between different characteristics variables of the participants.
Multivariate regression tree methodology was used for cluster analysis of bacterial abundance where clusters were age stratified. This analysis enabled us to determine the age limit with the highest explanatory power.
The Wilcoxon rank-sum test were performed on abundance data to explore the taxa significantly different among groups. The discriminatory power of a taxon was further assessed by calculating the area under receiver-operating characteristic curve (AUC) of a logistic regression model.
Acknowledgements
Not applicable.
Funding
The National Natural Science Foundation of China (No. 81573834), the Foundation of Key Laboratory of Ethnomedicine (Minzu University of China), Ministry of Education (No. KLEM-ZZ201808, KLEM-ZZ201802).
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the SRA database repository, Accession No: PRJNA533879.
Abbreviations
- ANOVA
One-way analysis of variance
- AUC
Area under receiver-operating characteristic curve
- CHO
Serum total cholesterol
- DM
Diabetic group
- FDR
False discovery rate
- GLU
Blood glucose level
- HDL-C
High-density lipoprotein cholesterol
- HGR
High blood glucose group
- IBD
Inflammatory bowel disease
- IDDM
Insulin-dependent diabetes mellitus
- IFG
Impaired fasting glucose group
- LDL-C
Low-density lipoprotein cholesterol
- LPS
Lipopolysaccharides
- MLG
Metagenomic linkage group
- MRT
Multivariate regress trees
- MUC
Minzu University of China
- NGR
Normal blood glucose group
- OTUs
Operational taxonomic units
- SCFAs
Short-chain fatty acids
- SD
Standard deviation
- T2D
Type 2 diabetes
- TG
Triglycerides
- TID
Type I diabetes
Authors’ contributions
SG, LS designed this work and revised the manuscript. WE, ZH developed the methodology analyzed and interpreted the data, WB, LS, XD acquired and managed patients, provided facilities. WR, LH conducted experiment. WE, ZH, WR wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was conducted with approval from the Ethics Committee of Minzu University of China (ECMUC2019003CO). All participants were enrolled in the study after obtaining both written and verbal consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wu Enqi, Email: w1n2q3@163.com.
Zhao Huanhu, Email: zhaohuanhu@163.com.
Wu Ritu, Email: wuritu0919@163.com.
Xie Dan, Email: 18811597518@163.com.
Lin Han, Email: lin_hanlh@163.com.
Wang Baili, Email: wangbaili@163.com.
Shen Gangyi, Phone: 86-10-6893-3254, Email: sgy@iccas.ac.cn.
Li Shuchun, Phone: 86-10-6893-3254, Email: jason@muc.edu.cn.
References
- 1.Leibson CL, Narayan KM. Trends in cardiovascular complications of diabetes. JAMA. 2005;293:1723. doi: 10.1001/jama.293.14.1723-a. [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee S, Khunti K, Davies MJ. Type 2 diabetes. Lancet. 2017;389:2239–2251. doi: 10.1016/S0140-6736(17)30058-2. [DOI] [PubMed] [Google Scholar]
- 3.Noecker C, McNally CP, Eng A, Borenstein E. High-resolution characterization of the human microbiome. Transl Res. 2017;179:7–23. doi: 10.1016/j.trsl.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prawitt J, Caron S, Staels B. Bile acid metabolism and the pathogenesis of type 2 diabetes. Curr Diab Rep. 2011;11:160–166. doi: 10.1007/s11892-011-0187-x). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J, Schoonjans K. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T, Chiang JY. Bile acids as metabolic regulators. Curr Opin Gastroenterol. 2015;31:159–165. doi: 10.1097/MOG.0000000000000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Toole Paul W., Jeffery Ian B. Microbiome–health interactions in older people. Cellular and Molecular Life Sciences. 2017;75(1):119–128. doi: 10.1007/s00018-017-2673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20:145–155. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YJ, Li S, Gan RY, Zhou T, Xu DP, Li HB. Impacts of gut bacteria on human health and diseases. Int J Mol Sci. 2015;16:7493–7519. doi: 10.3390/ijms16047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Membrez M, Blancher F, Jaquet M, Bibiloni R, Cani PD, Burcelin RG, Corthesy I, Mace K, Chou CJ. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008;22:2416–2426. doi: 10.1096/fj.07-102723. [DOI] [PubMed] [Google Scholar]
- 12.Diamant M, Blaak EE, de Vos WM. Do nutrient-gut-microbiota interactions play a role in human obesity, insulin resistance and type 2 diabetes? Obes Rev. 2011;12:272–281. doi: 10.1111/j.1467-789X.2010.00797.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Luo X, Mao X, Tao Y, Ran X, Zhao H, Xiong J, Li L. Gut microbiome analysis of type 2 diabetic patients from the Chinese minority ethnic groups the Uygurs and Kazaks. PLoS One. 2017;12:e0172774. doi: 10.1371/journal.pone.0172774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 15.Sepp E, Kolk H, Loivukene K, Mikelsaar M. Higher blood glucose level associated with body mass index and gut microbiota in elderly people. Microb Ecol Health Dis. 2014;25. 10.3402/mehd.v25.22857. [DOI] [PMC free article] [PubMed]
- 16.Bhute SS, Suryavanshi MV, Joshi SM, Yajnik CS, Shouche YS, Ghaskadbi SS. Gut microbial diversity assessment of Indian Type-2-diabetics reveals alterations in eubacteria, archaea, and eukaryotes. Front Microbiol. 2017;8:214. doi: 10.3389/fmicb.2017.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, Chen Y, Ji L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One. 2013;8:e71108. doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Ma C, Han L, Nawaz M, Gao F, Zhang X, Yu P, Zhao C, Li L, Zhou A, Wang J, Moore JE, Millar BC, Xu J. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr Microbiol. 2010;61:69–78. doi: 10.1007/s00284-010-9582-9. [DOI] [PubMed] [Google Scholar]
- 19.Sedighi M, Razavi S, Navab-Moghadam F, Khamseh ME, Alaei-Shahmiri F, Mehrtash A, Amirmozafari N. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb Pathog. 2017;111:362–369. doi: 10.1016/j.micpath.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, Nielsen J, Backhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 21.Tiihonen K, Ouwehand AC, Rautonen N. Human intestinal microbiota and healthy ageing. Ageing Res Rev. 2010;9:107–116. doi: 10.1016/j.arr.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Licastro F, Candore G, Lio D, Porcellini E, Colonna-Romano G, Franceschi C, Caruso C. Innate immunity and inflammation in ageing: a key for understanding age-related diseases. Immun Ageing. 2005;2:8. doi: 10.1186/1742-4933-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caruso C, Magrone T, Jirillo E. Low grade inflammation as a common pathogenetic denominator in age-related diseases: novel drug targets for anti-ageing strategies and successful ageing achievement. Part II. Curr Pharm Des. 2010;16:753. doi: 10.2174/138161210790883606. [DOI] [PubMed] [Google Scholar]
- 24.Buford TW. (Dis)trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome. 2017;5:80. doi: 10.1186/s40168-017-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen L, Duffy A. Factors influencing the gut microbiota, inflammation, and type 2 diabetes. J Nutr. 2017;147:1468S–1475S. doi: 10.3945/jn.116.240754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut. 2014;63:1513–1521. doi: 10.1136/gutjnl-2014-306928. [DOI] [PubMed] [Google Scholar]
- 27.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkila J, Monti D, Satokari R, Franceschi C, Brigidi P, De Vos W. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bian G, Gloor GB, Gong A, Jia C, Zhang W, Hu J, Zhang H, Zhang Y, Zhou Z, Zhang J, Burton JP, Reid G, Xiao Y, Zeng Q, Yang K, Li J. The gut microbiota of healthy aged Chinese is similar to that of the healthy young. Msphere. 2017;2. 10.1128/mSphere.00327-17 published online EpubSep-Oct. [DOI] [PMC free article] [PubMed]
- 29.Kong F, Hua Y, Zeng B, Ning R, Li Y, Zhao J. Gut microbiota signatures of longevity. Curr Biol. 2016;26:R832–R833. doi: 10.1016/j.cub.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 30.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jorgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clement K, Dore J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, H. I. T. c. Meta. Bork P, Wang J, Ehrlich SD, Pedersen O. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 31.Zaneveld JR, McMinds R, Vega Thurber R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol. 2017;2:17121. doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- 32.Karjalainen KM, Knuuttila ML, Kaar ML. Salivary factors in children and adolescents with insulin-dependent diabetes mellitus. Pediatr Dent. 1996;18:306–311. [PubMed] [Google Scholar]
- 33.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, Osawa R. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O'Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O'Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the SRA database repository, Accession No: PRJNA533879.