Figure 1. ZCN8 underlies flowering time QTL qDTA8.
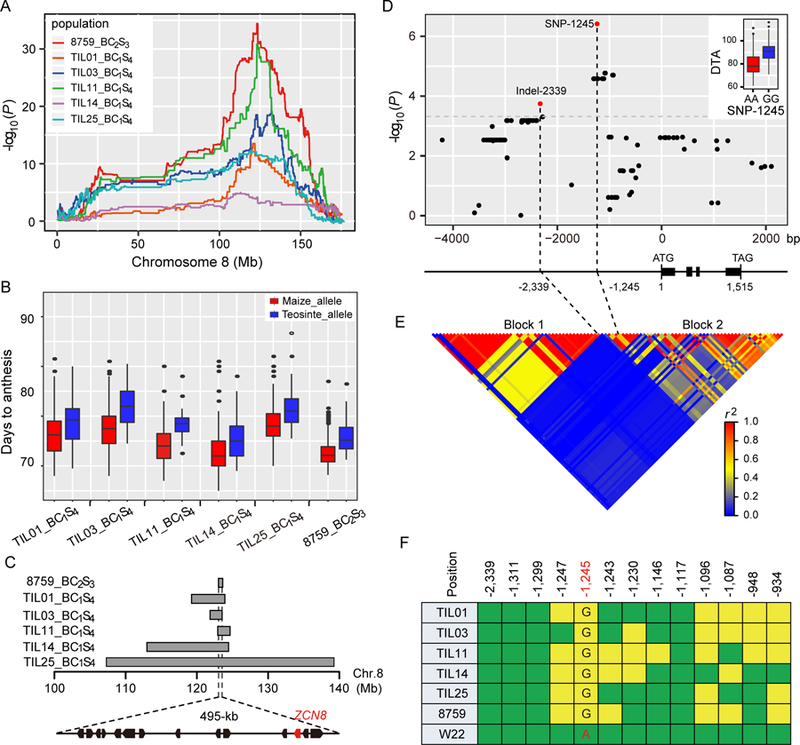
(A) qDTA8 was consistently mapped in six maize-teosinte experimental populations. The x-axis shows the physical position along chromosome 8, and the y-axis represents the logarithm of odds (LOD) score of each scanning position. Different coloured lines indicate different maize-teosinte populations. (B) Allele effects of qDTA8 in the six maize-teosinte populations. (C) ZCN8 is located in the overlapping region of qDTA8 in the six maize-teosinte populations. The arrows represent the 14 annotated genes in the reference genome in the overlapping region, and ZCN8 is indicated in red. (D) Association analysis of sequence variants in the 6.3-kb region of ZCN8 for DTA in a panel of 513 diverse maize inbred lines. SNP-1245 and Indel-2339, which are independently associated with DTA, are indicated by red dots. The gene structure of ZCN8 is shown below the association analysis plot. The positions of SNP-1245 and Indel-2339 relative to the start codon are indicated. The inset shows the difference in flowering time (days to anthesis) between the SNP-1245 alleles. (See also Table S1; Table S4 and Data S1). (E) Linkage disequilibrium (LD) analysis of the 6.3-kb region of ZCN8 in the maize association panel. The two LD blocks (Block 1 and Block 2) are indicated. (F) SNP-1245 co-segregates with qDTA8. The coloured squares indicate the allelic states of the six teosinte parents and the maize parent (W22) at the 13 significantly associated variants. The green squares indicate that the teosinte parents carry the same allele as W22, while the yellow squares indicate that the teosinte parents carry a different allele from W22. SNP-1245 is highlighted in red. (See also Table S2).
