Figure.
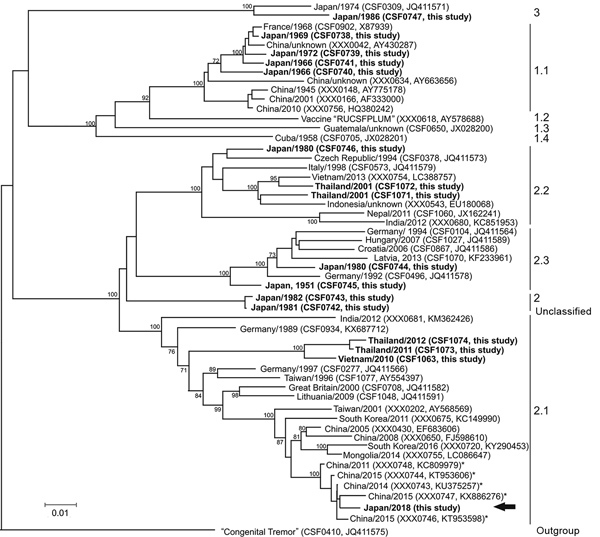
Phylogenetic tree displaying the genetic relatedness of the classical swine fever virus (CSFV) isolate obtained from the 2018 classical swine fever (CSF) outbreak in Japan to other CSFV isolates. Phylogenetic analyses were performed by using the neighbor-joining method with complete E2 (1,119 nt) sequences, including 1,000 iterations for bootstrap analysis, and were generated by the genetic typing module of the CSF database at the European Union and OIE Reference Laboratory for Classical Swine Fever (6). Only bootstrap values >70% are indicated. For each isolate, country and year are given, along with catalog number of the CSF database and the GenBank accession number in parentheses. The arrow indicates the sequence of the virus isolate obtained from a domestic pig during the 2018 CSF outbreak in Japan (Japan/2018) (GenBank accession no. LC425432). Boldface indicates additional newly generated sequences (GenBank accession nos. MK026451–65). Two ancient sequences from Japan (CSF0742, CSF0743) were identified as belonging to genotype 2 with no clear affiliation to any of the 3 established subgenotypes 2.1, 2.2, and 2.3 (indicated as genotype 2, unclassified). Asterisks indicate 5 E2 sequences belonging to a group of sequences that are most closely related to the E2 sequence of the CSFV isolate Japan/2018. Scale bar indicates nucleotide substitutions per site.
