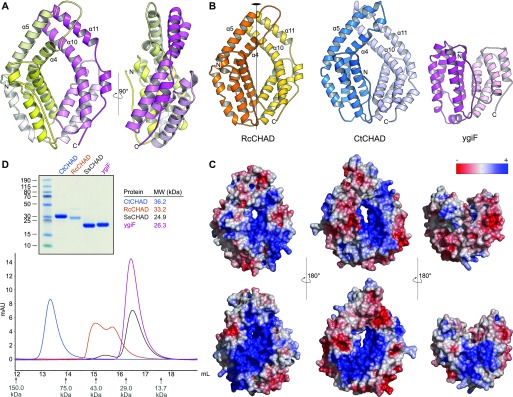
Figure 2. CHAD domains are helical bundles with twofold internal symmetry and a conserved basic surface area.
(A) Architecture of the CHAD domain. Shown is a ribbon diagram of RcCHAD colored from N- (yellow) to C-terminus (magenta). (B) Structural comparison of the RcCHAD, CtCHAD, and ygiF CHAD domain (Martinez et al, 2015) structures reveal the presence of two 4-helix bundles in all CHAD domains, related by pseudo twofold symmetry (indicated by a vertical line). Note that the helices α4, α6, α10, and α11 in ygiF are much shorter when compared with RcCHAD and CtCHAD, and hence ygiF lacks the central pore. (C) Identification of a conserved basic surface area in pro- and eukaryotic CHAD domains. Electrostatic potentials calculated in APBS (Jurrus et al, 2018) were mapped onto CHAD domain molecular surfaces in Pymol. Shown are front (upper panel) and back (lower panel) views. A highly basic surface area covers the front- and back side of the CHAD domain and includes the central pore present in RcCHAD and CtCHAD. (D) Analytical size-exclusion chromatography reveals different oligomeric states for the CHAD domains analyzed in this study. An SDS–PAGE analysis of the respective peak fractions (pooled) is shown alongside.