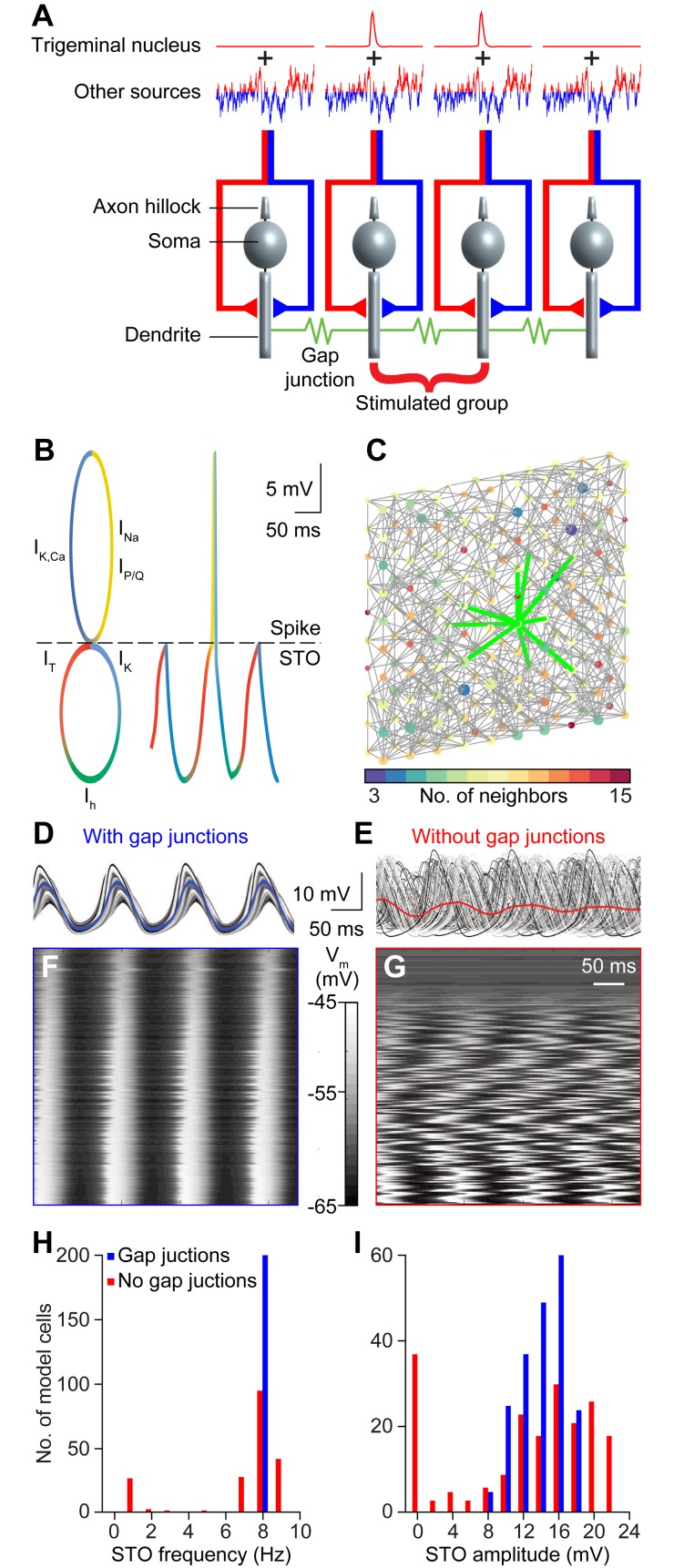
Fig 5. A tissue-scale network model of the inferior olive.
(A) To study the relative impact of the different components of the inferior olive we employed a biologically realistic network consisting of 200 model neurons embedded in a 3D grid of 10 x 10 x 2. The model neurons received input from two sources: a phasic input dubbed "sensory input" and a continuous fluctuating current, emulating a "contextual input". The “sensory input” reaches a subset of neurons synchronously and is modeled as an activation of glutamate receptor channels with a peak amplitude of 5 pA/cm2. The “sensory input” is complemented with a continuous input that is a mixture of excitatory and inhibitory synapses representing signals from different sources (see Fig 1A). The “contextual input” is modeled as an Ornstein-Uhlenbeck noise process delivered to all neurons in the model network, with a 20 ms decay, representing temporally correlated input. The model neurons are interconnected by gap junctions. (B) The model neurons display subthreshold membrane oscillations (STOs), represented by the lower circle, and spiking, represented by the upper circle. The colors indicate which current dominates which part of the activity pattern. An Ih current (green), a hyperpolarization-activated cation current, underlies a rise in membrane potential; the IT current (red), mediated by low threshold T-type Ca2+ channels (Cav3.1), further drives up the membrane potential and from here either the subthreshold oscillation continues (lower circle) or a spike is generated (upper circle), mediated through INa and IK currents (yellow and blue, respectively); both after a spike or a subthreshold peak, influx of Ca2+ through the P/Q-type type Ca2+ channels (Cav2.1) leads to a hyperpolarization due to Ca2+-dependent K+ channels (blue); this hyperpolarization in turn will activate the Ih current resuming the oscillation cycle. A resulting trace of the STO punctuated with one spike is diagrammatically displayed on the right. For a full description of currents, consult methods. (C) 3D connectivity scheme of the 200 neurons in the network model. Links indicate gap junctional interconnections; the color of each neuron in the model represents the number of neurons to which that cell is connected. The thick green lines indicate first order neighbors of the center cell. (D-E) Membrane potential of cells in the absence of input, for networks with and without gap junctional coupling. (F-G) Cell activity sorted by amplitude, indicating that coupling the neurons of the network brings non-oscillating cells into the synchronous oscillation. (H-I) Distribution of STO frequency and amplitude for coupled and uncoupled model networks.